Investigating the role of LSD2 as an epigenetic regulator in Ewing sarcoma
- PMID: 31231465
- PMCID: PMC6570473
- DOI: 10.18632/oncotarget.26988
Investigating the role of LSD2 as an epigenetic regulator in Ewing sarcoma
Abstract
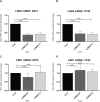
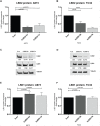
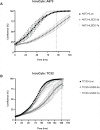
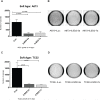
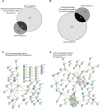
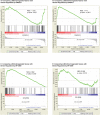
Ewing sarcoma is the second most common solid bone malignancy diagnosed in pediatric and young adolescent populations. Despite aggressive multi-modal treatment strategies, 5-year event-free survival remains at 75% for patients with localized disease and 20% for patients with metastases. Thus, the need for novel therapeutic options is imperative. Recent studies have focused on epigenetic misregulation in Ewing sarcoma development and potential new oncotargets for treatment. This project focused on the study of LSD2, a flavin-dependent histone demethylase found to be overexpressed in numerous cancers. We previously demonstrated that Ewing sarcoma cell lines are extremely susceptible to small molecule LSD1 blockade with SP-2509. Drug sensitivity correlated with the degree of LSD2 induction following treatment. As such, the purpose of this study was to determine the role of LSD2 in the epigenetic regulation of Ewing sarcoma, characterize genes regulated by LSD2, and examine the impact of SP-2509 drug treatment on LSD2 gene regulation. Genetic depletion (shRNA) of LSD2 significantly impaired oncogenic transformation with only a modest impact on proliferation. Transcriptional analysis of Ewing sarcoma cells following LSD2knockdown revealed modulation of genes primarily involved in metabolic regulation and nervous system development. Gene set enrichment analysis showed that SP-2509 does not impact LSD2 targeted genes. Although there are currently no small molecule agents that specifically target LSD2, our results support further investigations into agents that can inhibit this histone demethylase as a possible treatment for Ewing sarcoma.
Keywords: Ewing sarcoma; LSD1/KDM1A; LSD2/KDM1B; SP-2509.
Conflict of interest statement
CONFLICTS OF INTEREST Stephen L. Lessnick is a consultant/advisory board member for Salarius Pharmaceuticals.
Figures

References
-
- Pishas KI, Drenberg CD, Taslim C, Theisen ER, Johnson KM, Saund RS, Pop IL, Crompton BD, Lawlor ER, Tirode F, Mora J, Delattre O, Beckerle MC, et al.. Therapeutic Targeting of KDM1A/LSD1 in Ewing Sarcoma with SP-2509 Engages the Endoplasmic Reticulum Stress Response. Mol Cancer Ther. 2018; 17: 1902-16. 10.1158/1535-7163.MCT-18-0373. . - DOI - PMC - PubMed
-
- Sankar S, Theisen ER, Bearss J, Mulvihill T, Hoffman LM, Sorna V, Beckerle MC, Sharma S, Lessnick SL. Reversible LSD1 inhibition interferes with global EWS/ETS transcriptional activity and impedes Ewing sarcoma tumor growth. Clin Cancer Res. 2014; 20: 4584-97. 10.1158/1078-0432.CCR-14-0072. . - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

