Accelerated directed evolution of dye-decolorizing peroxidase using a bacterial extracellular protein secretion system (BENNY)
- PMID: 31231605
- PMCID: PMC6544594
- DOI: 10.1186/s40643-019-0255-7
Accelerated directed evolution of dye-decolorizing peroxidase using a bacterial extracellular protein secretion system (BENNY)
Abstract
Background: Dye-decolorizing peroxidases (DyPs) are haem-containing peroxidases that show great promises in industrial biocatalysis and lignocellulosic degradation. Through the use of Escherichia coli osmotically-inducible protein Y (OsmY) as a bacterial extracellular protein secretion system (BENNY), we successfully developed a streamlined directed evolution workflow to accelerate the protein engineering of DyP4 from Pleurotus ostreatus strain PC15.
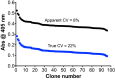
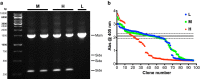
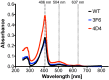
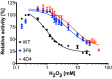
Result: After 3 rounds of random mutagenesis with error-prone polymerase chain reaction (epPCR) and 1 round of saturation mutagenesis, we obtained 4D4 variant (I56V, K109R, N227S and N312S) that displays multiple desirable phenotypes, including higher protein yield and secretion, higher specific activity (2.7-fold improvement in k cat/K m) and higher H2O2 tolerance (sevenfold improvement based on IC50).
Conclusion: To our best knowledge, this is the first report of applying OsmY to simplify the directed evolution workflow and to direct the extracellular secretion of a haem protein such as DyP4.
Keywords: Directed evolution; Dye-decolorizing peroxidase; Extracellular protein secretion; Hydrogen peroxide tolerance; Osmotically-inducible protein Y.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures

Similar articles
-
Description of the first fungal dye-decolorizing peroxidase oxidizing manganese(II).Appl Microbiol Biotechnol. 2015 Nov;99(21):8927-42. doi: 10.1007/s00253-015-6665-3. Epub 2015 May 13. Appl Microbiol Biotechnol. 2015. PMID: 25967658 Free PMC article.
-
Random and combinatorial mutagenesis for improved total production of secretory target protein in Escherichia coli.Sci Rep. 2021 Mar 5;11(1):5290. doi: 10.1038/s41598-021-84859-6. Sci Rep. 2021. PMID: 33674702 Free PMC article.
-
Comprehensive investigation of a dye-decolorizing peroxidase and a manganese peroxidase from Irpex lacteus F17, a lignin-degrading basidiomycete.AMB Express. 2018 Jul 17;8(1):119. doi: 10.1186/s13568-018-0648-6. AMB Express. 2018. PMID: 30019324 Free PMC article.
-
Biochemical features of dye-decolorizing peroxidases: Current impact on lignin degradation.Biotechnol Appl Biochem. 2020 Sep;67(5):751-759. doi: 10.1002/bab.2015. Epub 2020 Sep 13. Biotechnol Appl Biochem. 2020. PMID: 32860433 Review.
-
Basidiomycete DyPs: Genomic diversity, structural-functional aspects, reaction mechanism and environmental significance.Arch Biochem Biophys. 2015 May 15;574:66-74. doi: 10.1016/j.abb.2015.01.018. Epub 2015 Jan 28. Arch Biochem Biophys. 2015. PMID: 25637654 Review.
Cited by
-
Engineering Ag43 Signal Peptides with Bacterial Display and Selection.Methods Protoc. 2022 Dec 23;6(1):1. doi: 10.3390/mps6010001. Methods Protoc. 2022. PMID: 36648950 Free PMC article.
-
Mutational and structural analysis of an ancestral fungal dye-decolorizing peroxidase.FEBS J. 2021 Jun;288(11):3602-3618. doi: 10.1111/febs.15687. Epub 2021 Jan 8. FEBS J. 2021. PMID: 33369202 Free PMC article.
-
Hyperthermophilic xylanase and thermophilicity analysis by molecular dynamic simulation with quantum mechanics.Appl Microbiol Biotechnol. 2024 Dec 4;108(1):526. doi: 10.1007/s00253-024-13356-3. Appl Microbiol Biotechnol. 2024. PMID: 39630239 Free PMC article.
-
Challenges and Recent Advances in Enzyme-Mediated Wastewater Remediation-A Review.Nanomaterials (Basel). 2021 Nov 19;11(11):3124. doi: 10.3390/nano11113124. Nanomaterials (Basel). 2021. PMID: 34835887 Free PMC article. Review.
-
Computational Studies of Enzymes for C-F Bond Degradation and Functionalization.Chemphyschem. 2025 May 5;26(9):e202401130. doi: 10.1002/cphc.202401130. Epub 2025 Mar 6. Chemphyschem. 2025. PMID: 39962931 Free PMC article. Review.
References
-
- Brissos V, Tavares D, Sousa AC, Robalo MP, Martins LO. Engineering a bacterial DyP-type peroxidase for enhanced oxidation of lignin-related phenolics at alkaline pH. ACS Catal. 2017;7:3454–3465. doi: 10.1021/acscatal.6b03331. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

