TRAV1-2+ CD8+ T-cells including oligoconal expansions of MAIT cells are enriched in the airways in human tuberculosis
- PMID: 31231693
- PMCID: PMC6549148
- DOI: 10.1038/s42003-019-0442-2
TRAV1-2+ CD8+ T-cells including oligoconal expansions of MAIT cells are enriched in the airways in human tuberculosis
Abstract
Mucosal-associated invariant T (MAIT) cells typically express a TRAV1-2+ semi-invariant TCRα that enables recognition of bacterial, mycobacterial, and fungal riboflavin metabolites presented by MR1. MAIT cells are associated with immune control of bacterial and mycobacterial infections in murine models. Here, we report that a population of pro-inflammatory TRAV1-2+ CD8+ T cells are present in the airways and lungs of healthy individuals and are enriched in bronchoalveolar fluid of patients with active pulmonary tuberculosis (TB). High-throughput T cell receptor analysis reveals oligoclonal expansions of canonical and donor-unique TRAV1-2+ MAIT-consistent TCRα sequences within this population. Some of these cells demonstrate MR1-restricted mycobacterial reactivity and phenotypes suggestive of MAIT cell identity. These findings demonstrate enrichment of TRAV1-2+ CD8+ T cells with MAIT or MAIT-like features in the airways during active TB and suggest a role for these cells in the human pulmonary immune response to Mycobacterium tuberculosis.
Keywords: Mucosal immunology; T-cell receptor; Tuberculosis.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures



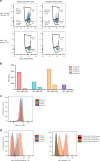
References
-
- Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J. Exp. Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007387/AI/NIAID NIH HHS/United States
- R01 AI048090/AI/NIAID NIH HHS/United States
- 107752/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- K08 AI118538/AI/NIAID NIH HHS/United States
- T32 HL083808/HL/NHLBI NIH HHS/United States
- R01 AI129980/AI/NIAID NIH HHS/United States
- R01 AI106725/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- P30 AI060354/AI/NIAID NIH HHS/United States
- R01 AI134790/AI/NIAID NIH HHS/United States
- I01 BX001231/BX/BLRD VA/United States
- R01 AI078965/AI/NIAID NIH HHS/United States
- R01 AI097138/AI/NIAID NIH HHS/United States
- R01 AI037856/AI/NIAID NIH HHS/United States
- I01 BX000533/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

