Multi-Institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation
- PMID: 31231720
- PMCID: PMC6589345
- DOI: 10.1007/978-3-030-11723-8_9
Multi-Institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation
Abstract
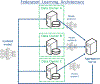
Deep learning models for semantic segmentation of images require large amounts of data. In the medical imaging domain, acquiring sufficient data is a significant challenge. Labeling medical image data requires expert knowledge. Collaboration between institutions could address this challenge, but sharing medical data to a centralized location faces various legal, privacy, technical, and data-ownership challenges, especially among international institutions. In this study, we introduce the first use of federated learning for multi-institutional collaboration, enabling deep learning modeling without sharing patient data. Our quantitative results demonstrate that the performance of federated semantic segmentation models (Dice=0.852) on multimodal brain scans is similar to that of models trained by sharing data (Dice=0.862). We compare federated learning with two alternative collaborative learning methods and find that they fail to match the performance of federated learning.
Keywords: BraTS; Deep Learning; Federated; Glioma; Incremental; Machine Learning; Segmentation.
Figures

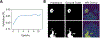
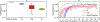
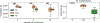
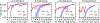
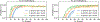
References
-
- Bakas S, Akbari H, Pisapia J, Martinez-Lage M, Rozycki M, Rathore S, Dahmane N, O’Rourke DM, Davatzikos C: In vivo detection of EGFRvIII in glioblastoma via perfusion magnetic resonance imaging signature consistent with deep peritumoral infiltration: the ∅-index. Clinical Cancer Research 23(16), 4724–4734 (2017) - PMC - PubMed
-
- Chang K, Bai HX, Zhou H, Su C, Bi WL, Agbodza E, Kavouridis VK, Senders JT, Boaro A, Beers A, Zhang B, Capellini A, Liao W, Shen Q, Li X, Xiao B, Cryan J, Ramkissoon S, Ramkissoon L, Ligon K, Wen PY, Bindra RS, Woo J, Arnaout O, Gerstner ER, Zhang PJ, Rosen BR, Yang L, Huang RY, Kalpathy-Cramer J: Residual Convolutional Neural Network for the Determination of ¡em¿IDH¡/em¿ Status in Low- and High-Grade Gliomas from MR Imaging. Clinical Cancer Research 24(5), 1073–1081 (2018) - PMC - PubMed
-
- Akbari H, Macyszyn L, Da X, Bilello M, Wolf RL, Martinez-Lage M, Biros G, Alonso-Basanta M, O’Rourke DM, Davatzikos C: Imaging Surrogates of Infiltration Obtained Via Multiparametric Imaging Pattern Analysis Predict Subsequent Location of Recurrence of Glioblastoma. Neurosurgery 78(4), 572–580 (2016) - PMC - PubMed
-
- Macyszyn L, Akbari H, Pisapia JM, Da X, Attiah M, Pigrish V, Bi Y, Pal S, Davuluri RV, Roccograndi L, Dahmane N, Martinez-Lage M, Biros G, Wolf RL, Bilello M, O’Rourke DM, Davatzikos C: Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro-Oncology 18(3), 417–425 (2016) - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
