Structural basis of Type IV CRISPR RNA biogenesis by a Cas6 endoribonuclease
- PMID: 31232162
- PMCID: PMC6779396
- DOI: 10.1080/15476286.2019.1634965
Structural basis of Type IV CRISPR RNA biogenesis by a Cas6 endoribonuclease
Abstract
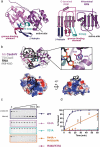
Prokaryotic CRISPR-Cas adaptive immune systems rely on small non-coding RNAs derived from CRISPR loci to recognize and destroy complementary nucleic acids. However, the mechanism of Type IV CRISPR RNA (crRNA) biogenesis is poorly understood. To dissect the mechanism of Type IV CRISPR RNA biogenesis, we determined the x-ray crystal structure of the putative Type IV CRISPR associated endoribonuclease Cas6 from Mahella australiensis (Ma Cas6-IV) and characterized its enzymatic activity with RNA cleavage assays. We show that Ma Cas6-IV specifically cleaves Type IV crRNA repeats at the 3' side of a predicted stem loop, with a metal-independent, single-turnover mechanism that relies on a histidine and a tyrosine located within the putative endonuclease active site. Structure and sequence alignments with Cas6 orthologs reveal that although Ma Cas6-IV shares little sequence homology with other Cas6 proteins, all share common structural features that bind distinct crRNA repeat sequences. This analysis of Type IV crRNA biogenesis provides a structural and biochemical framework for understanding the similarities and differences of crRNA biogenesis across multi-subunit Class 1 CRISPR immune systems.
Keywords: CRISPR; Cas6; Csf5; RNA; Type IV CRISPR; X-ray crystallography; endonuclease; prokaryotic immunity.
Figures



Similar articles
-
The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA.Nature. 2016 Apr 28;532(7600):517-21. doi: 10.1038/nature17945. Epub 2016 Apr 20. Nature. 2016. PMID: 27096362
-
Primary processing of CRISPR RNA by the endonuclease Cas6 in Staphylococcus epidermidis.FEBS Lett. 2015 Oct 7;589(20 Pt B):3197-204. doi: 10.1016/j.febslet.2015.09.005. Epub 2015 Sep 10. FEBS Lett. 2015. PMID: 26364721 Free PMC article.
-
Cutting it close: CRISPR-associated endoribonuclease structure and function.Trends Biochem Sci. 2015 Jan;40(1):58-66. doi: 10.1016/j.tibs.2014.10.007. Epub 2014 Nov 18. Trends Biochem Sci. 2015. PMID: 25468820 Review.
-
Cas6 specificity and CRISPR RNA loading in a complex CRISPR-Cas system.Nucleic Acids Res. 2014 Jun;42(10):6532-41. doi: 10.1093/nar/gku308. Epub 2014 Apr 20. Nucleic Acids Res. 2014. PMID: 24753403 Free PMC article.
-
Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR-Cas adaptive immunity.FEMS Microbiol Rev. 2015 May;39(3):428-41. doi: 10.1093/femsre/fuv023. Epub 2015 May 19. FEMS Microbiol Rev. 2015. PMID: 25994611 Free PMC article. Review.
Cited by
-
Identification of a Type IV-A CRISPR-Cas System Located Exclusively on IncHI1B/IncFIB Plasmids in Enterobacteriaceae.Front Microbiol. 2020 Aug 12;11:1937. doi: 10.3389/fmicb.2020.01937. eCollection 2020. Front Microbiol. 2020. PMID: 32903441 Free PMC article.
-
Evolution of Type IV CRISPR-Cas Systems: Insights from CRISPR Loci in Integrative Conjugative Elements of Acidithiobacillia.CRISPR J. 2021 Oct;4(5):656-672. doi: 10.1089/crispr.2021.0051. Epub 2021 Sep 28. CRISPR J. 2021. PMID: 34582696 Free PMC article.
-
Positioning Diverse Type IV Structures and Functions Within Class 1 CRISPR-Cas Systems.Front Microbiol. 2021 May 21;12:671522. doi: 10.3389/fmicb.2021.671522. eCollection 2021. Front Microbiol. 2021. PMID: 34093491 Free PMC article.
-
The arms race between bacteria and their phage foes.Nature. 2020 Jan;577(7790):327-336. doi: 10.1038/s41586-019-1894-8. Epub 2020 Jan 15. Nature. 2020. PMID: 31942051 Review.
-
Endogenous Type I CRISPR-Cas: From Foreign DNA Defense to Prokaryotic Engineering.Front Bioeng Biotechnol. 2020 Mar 4;8:62. doi: 10.3389/fbioe.2020.00062. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32195227 Free PMC article. Review.
References
-
- Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: harnessing Nature’s toolbox for genome engineering. Cell. 2016;164:29–44. - PubMed
-
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. - PubMed
-
- Mohanraju P, Makarova KS, Zetsche B, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
