Glycosylphosphatidylinositol biosynthesis and remodeling are required for neural tube closure, heart development, and cranial neural crest cell survival
- PMID: 31232685
- PMCID: PMC6611694
- DOI: 10.7554/eLife.45248
Glycosylphosphatidylinositol biosynthesis and remodeling are required for neural tube closure, heart development, and cranial neural crest cell survival
Abstract
Glycosylphosphatidylinositol (GPI) anchors attach nearly 150 proteins to the cell membrane. Patients with pathogenic variants in GPI biosynthesis genes develop diverse phenotypes including seizures, dysmorphic facial features and cleft palate through an unknown mechanism. We identified a novel mouse mutant (cleft lip/palate, edema and exencephaly; Clpex) with a hypo-morphic mutation in Post-Glycophosphatidylinositol Attachment to Proteins-2 (Pgap2), a component of the GPI biosynthesis pathway. The Clpex mutation decreases surface GPI expression. Surprisingly, Pgap2 showed tissue-specific expression with enrichment in the brain and face. We found the Clpex phenotype is due to apoptosis of neural crest cells (NCCs) and the cranial neuroepithelium. We showed folinic acid supplementation in utero can partially rescue the cleft lip phenotype. Finally, we generated a novel mouse model of NCC-specific total GPI deficiency. These mutants developed median cleft lip and palate demonstrating a previously undocumented cell autonomous role for GPI biosynthesis in NCC development.
Keywords: Folic/folinic acid; congenital disorders of glycosylation; craniofacial; development; developmental biology; glycosylphosphatidylinositol; mouse; neural crest.
© 2019, Lukacs et al.
Conflict of interest statement
ML, TR, PC, RS No competing interests declared
Figures








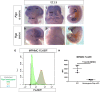
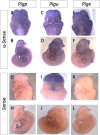
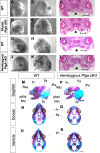
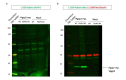
Similar articles
-
A post glycosylphosphatidylinositol (GPI) attachment to proteins, type 2 (PGAP2) variant identified in Mabry syndrome index cases: Molecular genetics of the prototypical inherited GPI disorder.Eur J Med Genet. 2020 Apr;63(4):103822. doi: 10.1016/j.ejmg.2019.103822. Epub 2019 Dec 2. Eur J Med Genet. 2020. PMID: 31805394
-
Eif3ba regulates cranial neural crest development by modulating p53 in zebrafish.Dev Biol. 2013 Sep 1;381(1):83-96. doi: 10.1016/j.ydbio.2013.06.009. Epub 2013 Jun 17. Dev Biol. 2013. PMID: 23791820
-
Significantly different clinical phenotypes associated with mutations in synthesis and transamidase+remodeling glycosylphosphatidylinositol (GPI)-anchor biosynthesis genes.Orphanet J Rare Dis. 2020 Feb 4;15(1):40. doi: 10.1186/s13023-020-1313-0. Orphanet J Rare Dis. 2020. PMID: 32019583 Free PMC article.
-
Cellular and molecular mechanisms in the development of a cleft lip and/or cleft palate; insights from zebrafish (Danio rerio).Anat Rec (Hoboken). 2021 Aug;304(8):1650-1660. doi: 10.1002/ar.24547. Epub 2020 Nov 2. Anat Rec (Hoboken). 2021. PMID: 33099891 Review.
-
CHO glycosylation mutants: GPI anchor.Methods Enzymol. 2006;416:182-205. doi: 10.1016/S0076-6879(06)16012-7. Methods Enzymol. 2006. PMID: 17113867 Review.
Cited by
-
Genome-wide DNA methylation profiling in blood reveals epigenetic signature of incident acute coronary syndrome.Nat Commun. 2024 Aug 28;15(1):7431. doi: 10.1038/s41467-024-51751-6. Nat Commun. 2024. PMID: 39198424 Free PMC article.
-
Limb-clasping, cognitive deficit and increased vulnerability to kainic acid-induced seizures in neuronal glycosylphosphatidylinositol deficiency mouse models.Hum Mol Genet. 2021 May 28;30(9):758-770. doi: 10.1093/hmg/ddab052. Hum Mol Genet. 2021. PMID: 33607654 Free PMC article.
-
CNS glycosylphosphatidylinositol deficiency results in delayed white matter development, ataxia and premature death in a novel mouse model.Hum Mol Genet. 2020 May 8;29(7):1205-1217. doi: 10.1093/hmg/ddaa046. Hum Mol Genet. 2020. PMID: 32179897 Free PMC article.
-
Novel investigations in retinoic-acid-induced cleft palate about the gut microbiome of pregnant mice.Front Cell Infect Microbiol. 2022 Dec 15;12:1042779. doi: 10.3389/fcimb.2022.1042779. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36590585 Free PMC article.
-
Genomic, transcriptomic, and metabolomic profiles of hiPSC-derived dopamine neurons from clinically discordant brothers with identical PRKN deletions.NPJ Parkinsons Dis. 2022 Jun 29;8(1):84. doi: 10.1038/s41531-022-00346-3. NPJ Parkinsons Dis. 2022. PMID: 35768426 Free PMC article.
References
-
- Bassuk AG, Muthuswamy LB, Boland R, Smith TL, Hulstrand AM, Northrup H, Hakeman M, Dierdorff JM, Yung CK, Long A, Brouillette RB, Au KS, Gurnett C, Houston DW, Cornell RA, Manak JR. Copy number variation analysis implicates the cell polarity gene glypican 5 as a human spina bifida candidate gene. Human Molecular Genetics. 2013;22:1097–1111. doi: 10.1093/hmg/dds515. - DOI - PMC - PubMed
-
- Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. Fourth edition. New York: Cold Spring Harbor Laboratory Press; 2014.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

