Improved CUT&RUN chromatin profiling tools
- PMID: 31232687
- PMCID: PMC6598765
- DOI: 10.7554/eLife.46314
Improved CUT&RUN chromatin profiling tools
Abstract
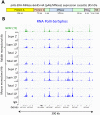
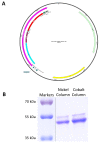
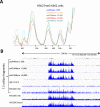
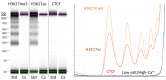
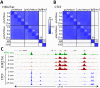
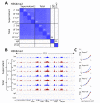
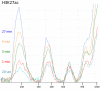
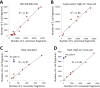
Previously, we described a novel alternative to chromatin immunoprecipitation, CUT&RUN, in which unfixed permeabilized cells are incubated with antibody, followed by binding of a protein A-Micrococcal Nuclease (pA/MNase) fusion protein (Skene and Henikoff, 2017). Here we introduce three enhancements to CUT&RUN: A hybrid protein A-Protein G-MNase construct that expands antibody compatibility and simplifies purification, a modified digestion protocol that inhibits premature release of the nuclease-bound complex, and a calibration strategy based on carry-over of E. coli DNA introduced with the fusion protein. These new features, coupled with the previously described low-cost, high efficiency, high reproducibility and high-throughput capability of CUT&RUN make it the method of choice for routine epigenomic profiling.
Keywords: chromatin; chromosomes; epigenomics; gene expression; genetics; genomics; human; spike-in calibration.
© 2019, Meers et al.
Conflict of interest statement
MM, TB, JH, SH No competing interests declared
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

