Comparative Analyses of Chromatin Landscape in White Adipose Tissue Suggest Humans May Have Less Beigeing Potential than Other Primates
- PMID: 31233101
- PMCID: PMC6648876
- DOI: 10.1093/gbe/evz134
Comparative Analyses of Chromatin Landscape in White Adipose Tissue Suggest Humans May Have Less Beigeing Potential than Other Primates
Abstract
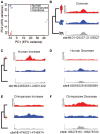
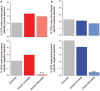
Humans carry a much larger percentage of body fat than other primates. Despite the central role of adipose tissue in metabolism, little is known about the evolution of white adipose tissue in primates. Phenotypic divergence is often caused by genetic divergence in cis-regulatory regions. We examined the cis-regulatory landscape of fat during human origins by performing comparative analyses of chromatin accessibility in human and chimpanzee adipose tissue using rhesus macaque as an outgroup. We find that many regions that have decreased accessibility in humans are enriched for promoter and enhancer sequences, are depleted for signatures of negative selection, are located near genes involved with lipid metabolism, and contain a short sequence motif involved in the beigeing of fat, the process in which lipid-storing white adipocytes are transdifferentiated into thermogenic beige adipocytes. The collective closing of many putative regulatory regions associated with beigeing of fat suggests a mechanism that increases body fat in humans.
Keywords: adipose; chromatin accessibility; comparative genomics; primates.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Intermittent fasting promotes type 3 innate lymphoid cells secreting IL-22 contributing to the beigeing of white adipose tissue.Elife. 2024 Mar 27;12:RP91060. doi: 10.7554/eLife.91060. Elife. 2024. PMID: 38536726 Free PMC article.
-
Effects of growth hormone on uncoupling protein 1 in white adipose tissues in obese mice.Growth Horm IGF Res. 2017 Dec;37:31-39. doi: 10.1016/j.ghir.2017.10.006. Epub 2017 Oct 24. Growth Horm IGF Res. 2017. PMID: 29111497
-
Adipocyte-specific Hypoxia-inducible gene 2 promotes fat deposition and diet-induced insulin resistance.Mol Metab. 2016 Sep 28;5(12):1149-1161. doi: 10.1016/j.molmet.2016.09.009. eCollection 2016 Dec. Mol Metab. 2016. PMID: 27900258 Free PMC article.
-
[Brown, white, beige: the color of fat and new therapeutic perspectives for obesity...].Ann Endocrinol (Paris). 2012 Oct;73 Suppl 1:S2-8. doi: 10.1016/S0003-4266(12)70009-4. Ann Endocrinol (Paris). 2012. PMID: 23089378 Review. French.
-
Metabolic interplay between white, beige, brown adipocytes and the liver.J Hepatol. 2016 May;64(5):1176-1186. doi: 10.1016/j.jhep.2016.01.025. Epub 2016 Jan 30. J Hepatol. 2016. PMID: 26829204 Review.
Cited by
-
Hybrid Epigenomes Reveal Extensive Local Genetic Changes to Chromatin Accessibility Contribute to Divergence in Embryonic Gene Expression Between Species.Mol Biol Evol. 2023 Nov 3;40(11):msad222. doi: 10.1093/molbev/msad222. Mol Biol Evol. 2023. PMID: 37823438 Free PMC article.
-
Systematic functional characterization of non-coding regulatory SNPs associated with central obesity.Am J Hum Genet. 2025 Jan 2;112(1):116-134. doi: 10.1016/j.ajhg.2024.11.005. Am J Hum Genet. 2025. PMID: 39753113 Free PMC article.
-
LINE retrotransposons characterize mammalian tissue-specific and evolutionarily dynamic regulatory regions.Genome Biol. 2021 Feb 18;22(1):62. doi: 10.1186/s13059-021-02260-y. Genome Biol. 2021. PMID: 33602314 Free PMC article.
-
Integrating the Thrifty Genotype and Evolutionary Mismatch Hypotheses to understand variation in cardiometabolic disease risk.Evol Med Public Health. 2024 Jul 31;12(1):214-226. doi: 10.1093/emph/eoae014. eCollection 2024. Evol Med Public Health. 2024. PMID: 39484023 Free PMC article. Review.
-
Chromatin activity identifies differential gene regulation across human ancestries.Genome Biol. 2024 Jan 15;25(1):21. doi: 10.1186/s13059-024-03165-2. Genome Biol. 2024. PMID: 38225662 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

