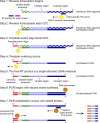
A simplified workflow for monoclonal antibody sequencing
- PMID: 31233538
- PMCID: PMC6590890
- DOI: 10.1371/journal.pone.0218717
A simplified workflow for monoclonal antibody sequencing
Abstract
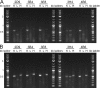
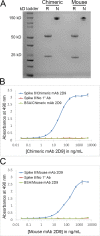
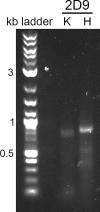
The diversity of antibody variable regions makes cDNA sequencing challenging, and conventional monoclonal antibody cDNA amplification requires the use of degenerate primers. Here, we describe a simplified workflow for amplification of IgG antibody variable regions from hybridoma RNA by a specialized RT-PCR followed by Sanger sequencing. We perform three separate reactions for each hybridoma: one each for kappa, lambda, and heavy chain transcripts. We prime reverse transcription with a primer specific to the respective constant region and use a template-switch oligonucleotide, which creates a custom sequence at the 5' end of the antibody cDNA. This template-switching circumvents the issue of low sequence homology and the need for degenerate primers. Instead, subsequent PCR amplification of the antibody cDNA molecules requires only two primers: one primer specific for the template-switch oligonucleotide sequence and a nested primer to the respective constant region. We successfully sequenced the variable regions of five mouse monoclonal IgG antibodies using this method, which enabled us to design chimeric mouse/human antibody expression plasmids for recombinant antibody production in mammalian cell culture expression systems. All five recombinant antibodies bind their respective antigens with high affinity, confirming that the amino acid sequences determined by our method are correct and demonstrating the high success rate of our method. Furthermore, we also designed RT-PCR primers and amplified the variable regions from RNA of cells transfected with chimeric mouse/human antibody expression plasmids, showing that our approach is also applicable to IgG antibodies of human origin. Our monoclonal antibody sequencing method is highly accurate, user-friendly, and very cost-effective.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Coco-Martin JM, Oberink JW, van der Velden-de Groot TAM, Beuvery EC. Methods for studying the stability of antibody expression by hybridoma cells in homogeneous continuous culture systems. Analytica Chimica Acta. 1991;249(1):257–62. 10.1016/0003-2670(91)87031-2. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

