Genome-Wide Analysis of Long Non-Coding RNA Profiles in Canine Oral Melanomas
- PMID: 31234577
- PMCID: PMC6628375
- DOI: 10.3390/genes10060477
Genome-Wide Analysis of Long Non-Coding RNA Profiles in Canine Oral Melanomas
Abstract
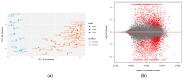
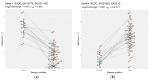
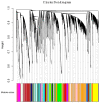
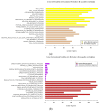
Mucosal melanomas (MM) are rare aggressive cancers in humans, and one of the most common forms of oral cancers in dogs. Similar biological and histological features are shared between MM in both species, making dogs a powerful model for comparative oncology studies of melanomas. Although exome sequencing recently identified recurrent coding mutations in canine MM, little is known about changes in non-coding gene expression, and more particularly, in canine long non-coding RNAs (lncRNAs), which are commonly dysregulated in human cancers. Here, we sampled a large cohort (n = 52) of canine normal/tumor oral MM from three predisposed breeds (poodles, Labrador retrievers, and golden retrievers), and used deep transcriptome sequencing to identify more than 400 differentially expressed (DE) lncRNAs. We further prioritized candidate lncRNAs by comparative genomic analysis to pinpoint 26 dog-human conserved DE lncRNAs, including SOX21-AS, ZEB2-AS, and CASC15 lncRNAs. Using unsupervised co-expression network analysis with coding genes, we inferred the potential functions of the DE lncRNAs, suggesting associations with cancer-related genes, cell cycle, and carbohydrate metabolism Gene Ontology (GO) terms. Finally, we exploited our multi-breed design to identify DE lncRNAs within breeds. This study provides a unique transcriptomic resource for studying oral melanoma in dogs, and highlights lncRNAs that may potentially be diagnostic or therapeutic targets for human and veterinary medicine.
Keywords: dogs; long non-coding RNAs (lncRNAs); mucosal melanoma; transcriptome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
A comprehensive genome-wide analysis of the long noncoding RNA expression profile in metastatic lymph nodes of oral mucosal melanoma.Gene. 2018 Oct 30;675:44-53. doi: 10.1016/j.gene.2018.06.064. Epub 2018 Jun 28. Gene. 2018. PMID: 29960071
-
Canine Melanomas as Models for Human Melanomas: Clinical, Histological, and Genetic Comparison.Genes (Basel). 2019 Jun 30;10(7):501. doi: 10.3390/genes10070501. Genes (Basel). 2019. PMID: 31262050 Free PMC article. Review.
-
Identification of mutations in canine oral mucosal melanomas by exome sequencing and comparison with human melanomas.Sci Rep. 2024 Oct 15;14(1):24174. doi: 10.1038/s41598-024-74748-z. Sci Rep. 2024. PMID: 39406779 Free PMC article.
-
Transcriptome sequencing reveals thousands of novel long non-coding RNAs in B cell lymphoma.Genome Med. 2015 Nov 1;7:110. doi: 10.1186/s13073-015-0230-7. Genome Med. 2015. PMID: 26521025 Free PMC article.
-
Long non-coding RNAs: implications in targeted diagnoses, prognosis, and improved therapeutic strategies in human non- and triple-negative breast cancer.Clin Epigenetics. 2018 Jun 27;10:88. doi: 10.1186/s13148-018-0514-z. eCollection 2018. Clin Epigenetics. 2018. PMID: 29983835 Free PMC article. Review.
Cited by
-
Cross-species analysis of enhancer logic using deep learning.Genome Res. 2020 Dec;30(12):1815-1834. doi: 10.1101/gr.260844.120. Epub 2020 Jul 30. Genome Res. 2020. PMID: 32732264 Free PMC article.
-
A Comparative View on Molecular Alterations and Potential Therapeutic Strategies for Canine Oral Melanoma.Vet Sci. 2021 Nov 22;8(11):286. doi: 10.3390/vetsci8110286. Vet Sci. 2021. PMID: 34822659 Free PMC article. Review.
-
Long Non-Coding RNA as a Potential Biomarker for Canine Tumors.Vet Sci. 2023 Oct 30;10(11):637. doi: 10.3390/vetsci10110637. Vet Sci. 2023. PMID: 37999460 Free PMC article.
-
Spontaneously occurring melanoma in animals and their relevance to human melanoma.J Pathol. 2020 Sep;252(1):4-21. doi: 10.1002/path.5505. Epub 2020 Jul 31. J Pathol. 2020. PMID: 32652526 Free PMC article. Review.
-
Five genetic variants explain over 70% of hair coat pheomelanin intensity variation in purebred and mixed breed domestic dogs.PLoS One. 2021 May 27;16(5):e0250579. doi: 10.1371/journal.pone.0250579. eCollection 2021. PLoS One. 2021. PMID: 34043658 Free PMC article.
References
-
- Wong K., van der Weyden L., Schott C.R., Foote A., Constantino-Casas F., Smith S., Dobson J.M., Murchison E.P., Wu H., Yeh I., et al. Cross-species genomic landscape comparison of human mucosal melanoma with canine oral and equine melanoma. Nat. Commun. 2019;10:353. doi: 10.1038/s41467-018-08081-1. - DOI - PMC - PubMed
-
- Hendricks W.P.D., Zismann V., Sivaprakasam K., Legendre C., Poorman K., Tembe W., Perdigones N., Kiefer J., Liang W., DeLuca V., et al. Trent, Somatic inactivating PTPRJ mutations and dysregulated pathways identified in canine malignant melanoma by integrated comparative genomic analysis. PLoS Genet. 2018;14:e1007589. doi: 10.1371/journal.pgen.1007589. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

