Effect of a Russian-backbone live-attenuated influenza vaccine with an updated pandemic H1N1 strain on shedding and immunogenicity among children in The Gambia: an open-label, observational, phase 4 study
- PMID: 31235405
- PMCID: PMC6650545
- DOI: 10.1016/S2213-2600(19)30086-4
Effect of a Russian-backbone live-attenuated influenza vaccine with an updated pandemic H1N1 strain on shedding and immunogenicity among children in The Gambia: an open-label, observational, phase 4 study
Abstract
Background: The efficacy and effectiveness of the pandemic H1N1 (pH1N1) component in live attenuated influenza vaccine (LAIV) is poor. The reasons for this paucity are unclear but could be due to impaired replicative fitness of pH1N1 A/California/07/2009-like (Cal09) strains. We assessed whether an updated pH1N1 strain in the Russian-backbone trivalent LAIV resulted in greater shedding and immunogenicity compared with LAIV with Cal09.
Methods: We did an open-label, prospective, observational, phase 4 study in Sukuta, a periurban area in The Gambia. We enrolled children aged 24-59 months who were clinically well. Children received one dose of the WHO prequalified Russian-backbone trivalent LAIV containing either A/17/California/2009/38 (Cal09) or A/17/New York/15/5364 (NY15) based on their year of enrolment. Primary outcomes were the percentage of children with LAIV strain shedding at day 2 and day 7, haemagglutinin inhibition seroconversion, and an increase in influenza haemagglutinin-specific IgA and T-cell responses at day 21 after LAIV. This study is nested within a randomised controlled trial investigating LAIV-microbiome interactions (NCT02972957).
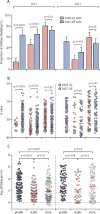
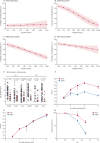
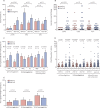
Findings: Between Feb 8, 2017, and April 12, 2017, 118 children were enrolled and received one dose of the Cal09 LAIV from 2016-17. Between Jan 15, 2018, and March 28, 2018, a separate cohort of 135 children were enrolled and received one dose of the NY15 LAIV from 2017-18, of whom 126 children completed the study. Cal09 showed impaired pH1N1 nasopharyngeal shedding (16 of 118 children [14%, 95% CI 8·0-21·1] with shedding at day 2 after administration of LAIV) compared with H3N2 (54 of 118 [46%, 36·6-55·2]; p<0·0001) and influenza B (95 of 118 [81%, 72·2-87·2]; p<0·0001), along with suboptimal serum antibody (seroconversion in six of 118 [5%, 1·9-10·7]) and T-cell responses (CD4+ interferon γ-positive and/or CD4+ interleukin 2-positive responses in 45 of 111 [41%, 31·3-50·3]). After the switch to NY15, a significant increase in pH1N1 shedding was seen (80 of 126 children [63%, 95% CI 54·4-71·9]; p<0·0001 compared with Cal09), along with improvements in seroconversion (24 of 126 [19%, 13·2-26·8]; p=0·011) and influenza-specific CD4+ T-cell responses (73 of 111 [66%, 60·0-75·6; p=0·00028]). The improvement in pH1N1 seroconversion with NY15 was even greater in children who were seronegative at baseline (24 of 64 children [38%, 95% CI 26·7-49·8] vs six of 79 children with Cal09 [8%, 2·8-15·8]; p<0·0001). Persistent shedding to day 7 was independently associated with both seroconversion (odds ratio 12·69, 95% CI 4·1-43·6; p<0·0001) and CD4+ T-cell responses (odds ratio 7·83, 95% CI 2·99-23·5; p<0·0001) by multivariable logistic regression.
Interpretation: The pH1N1 component switch that took place between 2016 and 2018 might have overcome the poor efficacy and effectiveness reported with previous LAIV formulations. LAIV effectiveness against pH1N1 should, therefore, improve in upcoming influenza seasons. Our data highlight the importance of assessing replicative fitness, in addition to antigenicity, when selecting annual LAIV components.
Funding: The Wellcome Trust.
Copyright © 2019 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY-NC-ND 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Figures
Comment in
-
Live attenuated influenza vaccines for African children.Lancet Respir Med. 2019 Aug;7(8):641-643. doi: 10.1016/S2213-2600(19)30145-6. Epub 2019 Jun 21. Lancet Respir Med. 2019. PMID: 31235406 No abstract available.
References
-
- Rhorer J, Ambrose CS, Dickinson S. Efficacy of live attenuated influenza vaccine in children: a meta-analysis of nine randomized clinical trials. Vaccine. 2009;27:1101–1110. - PubMed
-
- Flannery B, Chung J. Influenza vaccine effectiveness, including LAIV vs IIV in children and adolescents, US flu VE network, 2015–16. June 22, 2016. https://stacks.cdc.gov/view/cdc/60667
-
- Singanayagam A, Zambon M, Lalvani A, Barclay W. Urgent challenges in implementing live attenuated influenza vaccine. Lancet Infect Dis. 2018;18:e25–e32. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

