Mitochondrial fatty acid oxidation and the electron transport chain comprise a multifunctional mitochondrial protein complex
- PMID: 31235473
- PMCID: PMC6699831
- DOI: 10.1074/jbc.RA119.008680
Mitochondrial fatty acid oxidation and the electron transport chain comprise a multifunctional mitochondrial protein complex
Abstract
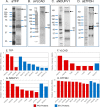
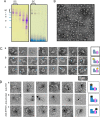
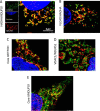
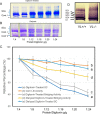
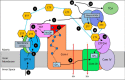
Three mitochondrial metabolic pathways are required for efficient energy production in eukaryotic cells: the electron transfer chain (ETC), fatty acid β-oxidation (FAO), and the tricarboxylic acid cycle. The ETC is organized into inner mitochondrial membrane supercomplexes that promote substrate channeling and catalytic efficiency. Although previous studies have suggested functional interaction between FAO and the ETC, their physical interaction has never been demonstrated. In this study, using blue native gel and two-dimensional electrophoreses, nano-LC-MS/MS, immunogold EM, and stimulated emission depletion microscopy, we show that FAO enzymes physically interact with ETC supercomplexes at two points. We found that the FAO trifunctional protein (TFP) interacts with the NADH-binding domain of complex I of the ETC, whereas the electron transfer enzyme flavoprotein dehydrogenase interacts with ETC complex III. Moreover, the FAO enzyme very-long-chain acyl-CoA dehydrogenase physically interacted with TFP, thereby creating a multifunctional energy protein complex. These findings provide a first view of an integrated molecular architecture for the major energy-generating pathways in mitochondria that ensures the safe transfer of unstable reducing equivalents from FAO to the ETC. They also offer insight into clinical ramifications for individuals with genetic defects in these pathways.
Keywords: electron microscopy (EM); fatty acid oxidation; mitochondrial metabolism; mitochondrial respiratory chain complex; primary metabolism; protein structure; proteomics; stimulated emission depletion microscopy (STED); supercomplex; trifunctional protein (TFP).
© 2019 Wang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Vockley J., Bennett M. J., and Gillingham M. B. (2016) in The Online Metabolic and Molecular Bases of Inherited Disease (Beaudet A. L., Vogelstein B., Kinzler K. W., Antonarakis S. E., Ballabio A., Gibson K. M., and Mitchell G., eds) McGraw-Hill Co., Inc., New York
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

