Guanine Quadruplex DNA Regulates Gamma Radiation Response of Genome Functions in the Radioresistant Bacterium Deinococcus radiodurans
- PMID: 31235513
- PMCID: PMC6689300
- DOI: 10.1128/JB.00154-19
Guanine Quadruplex DNA Regulates Gamma Radiation Response of Genome Functions in the Radioresistant Bacterium Deinococcus radiodurans
Abstract
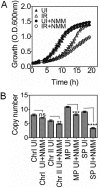
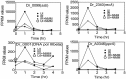
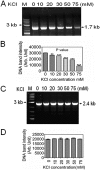
Guanine quadruplex (G4) DNA/RNA are secondary structures that regulate the various cellular processes in both eukaryotes and bacteria. Deinococcus radiodurans, a Gram-positive bacterium known for its extraordinary radioresistance, shows a genomewide occurrence of putative G4 DNA-forming motifs in its GC-rich genome. N-Methyl mesoporphyrin (NMM), a G4 DNA structure-stabilizing drug, did not affect bacterial growth under normal conditions but inhibited the postirradiation recovery of gamma-irradiated cells. Transcriptome sequencing analysis of cells treated with both radiation and NMM showed repression of gamma radiation-responsive gene expression, which was observed in the absence of NMM. Notably, this effect of NMM on the expression of housekeeping genes involved in other cellular processes was not observed. Stabilization of G4 DNA structures mapped at the upstream of recA and in the encoding region of DR_2199 had negatively affected promoter activity in vivo, DNA synthesis in vitro and protein translation in Escherichia coli host. These results suggested that G4 DNA plays an important role in DNA damage response and in the regulation of expression of the DNA repair proteins required for radioresistance in D. radioduransIMPORTANCEDeinococcus radiodurans can recover from extensive DNA damage caused by many genotoxic agents. It lacks LexA/RecA-mediated canonical SOS response. Therefore, the molecular mechanisms underlying the regulation of DNA damage response would be worth investigating in this bacterium. D. radiodurans genome is GC-rich and contains numerous islands of putative guanine quadruplex (G4) DNA structure-forming motifs. Here, we showed that in vivo stabilization of G4 DNA structures can impair DNA damage response processes in D. radiodurans Essential cellular processes such as transcription, DNA synthesis, and protein translation, which are also an integral part of the double-strand DNA break repair pathway, are affected by the arrest of G4 DNA structure dynamics. Thus, the role of DNA secondary structures in DNA damage response and radioresistance is demonstrated.
Keywords: DSB repair; Deinococcus; guanine quadruplex DNA; radioresistance; transcriptomics.
Copyright © 2019 American Society for Microbiology.
Figures




Similar articles
-
G-quadruplex forming structural motifs in the genome of Deinococcus radiodurans and their regulatory roles in promoter functions.Appl Microbiol Biotechnol. 2015 Nov;99(22):9761-9. doi: 10.1007/s00253-015-6808-6. Epub 2015 Jul 23. Appl Microbiol Biotechnol. 2015. PMID: 26201493
-
DrRecQ regulates guanine quadruplex DNA structure dynamics and its impact on radioresistance in Deinococcus radiodurans.Mol Microbiol. 2019 Sep;112(3):854-865. doi: 10.1111/mmi.14321. Epub 2019 Jun 19. Mol Microbiol. 2019. PMID: 31162841
-
Genome-wide study predicts promoter-G4 DNA motifs regulate selective functions in bacteria: radioresistance of D. radiodurans involves G4 DNA-mediated regulation.Nucleic Acids Res. 2013 Jan 7;41(1):76-89. doi: 10.1093/nar/gks1071. Epub 2012 Nov 17. Nucleic Acids Res. 2013. PMID: 23161683 Free PMC article.
-
G-quadruplex DNA structures and their relevance in radioprotection.Biochim Biophys Acta Gen Subj. 2021 May;1865(5):129857. doi: 10.1016/j.bbagen.2021.129857. Epub 2021 Jan 27. Biochim Biophys Acta Gen Subj. 2021. PMID: 33508382 Review.
-
Deinococcus radiodurans: what belongs to the survival kit?Crit Rev Biochem Mol Biol. 2008 May-Jun;43(3):221-38. doi: 10.1080/10409230802122274. Crit Rev Biochem Mol Biol. 2008. PMID: 18568848 Review.
Cited by
-
Never Cared for What They Do: High Structural Stability of Guanine-Quadruplexes in the Presence of Strand-Break Damage.Molecules. 2022 May 19;27(10):3256. doi: 10.3390/molecules27103256. Molecules. 2022. PMID: 35630732 Free PMC article.
-
DivIVA Phosphorylation Affects Its Dynamics and Cell Cycle in Radioresistant Deinococcus radiodurans.Microbiol Spectr. 2023 Feb 6;11(2):e0314122. doi: 10.1128/spectrum.03141-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36744915 Free PMC article.
-
FtsK, a DNA Motor Protein, Coordinates the Genome Segregation and Early Cell Division Processes in Deinococcus radiodurans.mBio. 2022 Dec 20;13(6):e0174222. doi: 10.1128/mbio.01742-22. Epub 2022 Oct 27. mBio. 2022. PMID: 36300930 Free PMC article.
-
DNA damage response and cell cycle regulation in bacteria: a twist around the paradigm.Front Microbiol. 2024 Mar 28;15:1389074. doi: 10.3389/fmicb.2024.1389074. eCollection 2024. Front Microbiol. 2024. PMID: 38605710 Free PMC article.
-
Emerging roles of i-motif in gene expression and disease treatment.Front Pharmacol. 2023 Mar 20;14:1136251. doi: 10.3389/fphar.2023.1136251. eCollection 2023. Front Pharmacol. 2023. PMID: 37021044 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

