Tick saliva protein Evasin-3 modulates chemotaxis by disrupting CXCL8 interactions with glycosaminoglycans and CXCR2
- PMID: 31235521
- PMCID: PMC6699855
- DOI: 10.1074/jbc.RA119.008902
Tick saliva protein Evasin-3 modulates chemotaxis by disrupting CXCL8 interactions with glycosaminoglycans and CXCR2
Abstract
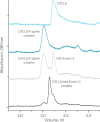
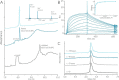
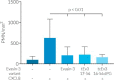
Chemokines are a group of chemotaxis proteins that regulate cell trafficking and play important roles in immune responses and inflammation. Ticks are blood-sucking parasites that secrete numerous immune-modulatory agents in their saliva to evade host immune responses. Evasin-3 is a small salivary protein that belongs to a class of chemokine-binding proteins isolated from the brown dog tick, Rhipicephalus sanguineus Evasin-3 has been shown to have a high affinity for chemokines CXCL1 and CXCL8 and to diminish inflammation in mice. In the present study, solution NMR spectroscopy was used to investigate the structure of Evasin-3 and its CXCL8-Evasin-3 complex. Evasin-3 is found to disrupt the glycosaminoglycan-binding site of CXCL8 and inhibit the interaction of CXCL8 with CXCR2. Structural data were used to design two novel CXCL8-binding peptides. The linear tEv3 17-56 and cyclic tcEv3 16-56 dPG Evasin-3 variants were chemically synthesized by solid-phase peptide synthesis. The affinity of these newly synthesized variants to CXCL8 was measured by surface plasmon resonance biosensor analysis. The Kd values of tEv3 17-56 and tcEv3 16-56 dPG were 27 and 13 nm, respectively. Both compounds effectively inhibited CXCL8-induced migration of polymorphonuclear neutrophils. The present results suggest utility of synthetic Evasin-3 variants as scaffolds for designing and fine-tuning new chemokine-binding agents that suppress immune responses and inflammation.
Keywords: C-X-C motif chemokine ligand (CXCL); chemokine; nuclear magnetic resonance (NMR); peptide chemical synthesis; protein structure; protein-protein interaction.
© 2019 Denisov et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
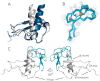
Similar articles
-
Structural characterization of anti-CCL5 activity of the tick salivary protein evasin-4.J Biol Chem. 2020 Oct 16;295(42):14367-14378. doi: 10.1074/jbc.RA120.013891. Epub 2020 Aug 14. J Biol Chem. 2020. PMID: 32817341 Free PMC article.
-
Molecular cloning and characterization of a highly selective chemokine-binding protein from the tick Rhipicephalus sanguineus.J Biol Chem. 2007 Sep 14;282(37):27250-27258. doi: 10.1074/jbc.M704706200. Epub 2007 Jul 19. J Biol Chem. 2007. PMID: 17640866
-
Tick Saliva Protein Evasin-3 Allows for Visualization of Inflammation in Arteries through Interactions with CXC-Type Chemokines Deposited on Activated Endothelium.Bioconjug Chem. 2020 Mar 18;31(3):948-955. doi: 10.1021/acs.bioconjchem.0c00095. Epub 2020 Mar 4. Bioconjug Chem. 2020. PMID: 32077689 Free PMC article.
-
Structural insights into the activation of chemokine receptor CXCR2.FEBS J. 2022 Jan;289(2):386-393. doi: 10.1111/febs.15865. Epub 2021 Apr 21. FEBS J. 2022. PMID: 33835690 Review.
-
Combined anti CXC receptors 1 and 2 therapy is a promising anti-inflammatory treatment for respiratory diseases by reducing neutrophil migration and activation.Pulm Pharmacol Ther. 2015 Oct;34:37-45. doi: 10.1016/j.pupt.2015.08.002. Epub 2015 Aug 10. Pulm Pharmacol Ther. 2015. PMID: 26271598 Review.
Cited by
-
Structural characterization of anti-CCL5 activity of the tick salivary protein evasin-4.J Biol Chem. 2020 Oct 16;295(42):14367-14378. doi: 10.1074/jbc.RA120.013891. Epub 2020 Aug 14. J Biol Chem. 2020. PMID: 32817341 Free PMC article.
-
Phylogenetic Analysis Indicates That Evasin-Like Proteins of Ixodid Ticks Fall Into Three Distinct Classes.Front Cell Infect Microbiol. 2021 Oct 22;11:769542. doi: 10.3389/fcimb.2021.769542. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34746035 Free PMC article.
-
Chemokine Heteromers and Their Impact on Cellular Function-A Conceptual Framework.Int J Mol Sci. 2023 Jun 30;24(13):10925. doi: 10.3390/ijms241310925. Int J Mol Sci. 2023. PMID: 37446102 Free PMC article. Review.
-
Binding Molecules in Tick Saliva for Targeting Host Cytokines, Chemokines, and Beyond.Biomolecules. 2024 Dec 21;14(12):1647. doi: 10.3390/biom14121647. Biomolecules. 2024. PMID: 39766354 Free PMC article. Review.
-
Malaria parasites both repress host CXCL10 and use it as a cue for growth acceleration.Nat Commun. 2021 Aug 11;12(1):4851. doi: 10.1038/s41467-021-24997-7. Nat Commun. 2021. PMID: 34381047 Free PMC article.
References
-
- Zheng Y., Han G. W., Abagyan R., Wu B., Stevens R. C., Cherezov V., Kufareva I., and Handel T. M. (2017) Structure of CC chemokine receptor 5 with a potent chemokine antagonist reveals mechanisms of chemokine recognition and molecular mimicry by HIV. Immunity 46, 1005–1017.e5 10.1016/j.immuni.2017.05.002 - DOI - PMC - PubMed
-
- Burg J. S., Ingram J. R., Venkatakrishnan A. J., Jude K. M., Dukkipati A., Feinberg E. N., Angelini A., Waghray D., Dror R. O., Ploegh H. L., and Garcia K. C. (2015) Structural biology: structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science 347, 1113–1117 10.1126/science.aaa5026 - DOI - PMC - PubMed
-
- Qin L., Kufareva I., Holden L. G., Wang C., Zheng Y., Zhao C., Fenalti G., Wu H., Han G. W., Cherezov V., Abagyan R., Stevens R. C., and Handel T. M. (2015) Structural biology. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science 347, 1117–1122 10.1126/science.1261064 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

