Enabling microbial syringol conversion through structure-guided protein engineering
- PMID: 31235604
- PMCID: PMC6628648
- DOI: 10.1073/pnas.1820001116
Enabling microbial syringol conversion through structure-guided protein engineering
Abstract
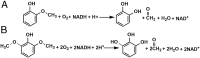
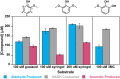
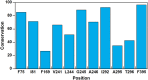
Microbial conversion of aromatic compounds is an emerging and promising strategy for valorization of the plant biopolymer lignin. A critical and often rate-limiting reaction in aromatic catabolism is O-aryl-demethylation of the abundant aromatic methoxy groups in lignin to form diols, which enables subsequent oxidative aromatic ring-opening. Recently, a cytochrome P450 system, GcoAB, was discovered to demethylate guaiacol (2-methoxyphenol), which can be produced from coniferyl alcohol-derived lignin, to form catechol. However, native GcoAB has minimal ability to demethylate syringol (2,6-dimethoxyphenol), the analogous compound that can be produced from sinapyl alcohol-derived lignin. Despite the abundance of sinapyl alcohol-based lignin in plants, no pathway for syringol catabolism has been reported to date. Here we used structure-guided protein engineering to enable microbial syringol utilization with GcoAB. Specifically, a phenylalanine residue (GcoA-F169) interferes with the binding of syringol in the active site, and on mutation to smaller amino acids, efficient syringol O-demethylation is achieved. Crystallography indicates that syringol adopts a productive binding pose in the variant, which molecular dynamics simulations trace to the elimination of steric clash between the highly flexible side chain of GcoA-F169 and the additional methoxy group of syringol. Finally, we demonstrate in vivo syringol turnover in Pseudomonas putida KT2440 with the GcoA-F169A variant. Taken together, our findings highlight the significant potential and plasticity of cytochrome P450 aromatic O-demethylases in the biological conversion of lignin-derived aromatic compounds.
Keywords: P450; biorefinery; demethylase; lignin.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
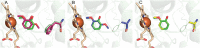

References
-
- Boerjan W., Ralph J., Baucher M., Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546 (2003). - PubMed
-
- Fuchs G., Boll M., Heider J., Microbial degradation of aromatic compounds—from one strategy to four. Nat. Rev. Microbiol. 9, 803–816 (2011). - PubMed
-
- Bugg T. D., Ahmad M., Hardiman E. M., Singh R., The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22, 394–400 (2011). - PubMed
-
- Floudas D., et al. , The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719 (2012). - PubMed
-
- Salvachúa D., et al. , Lignin depolymerization by fungal secretomes and a microbial sink. Green Chem. 18, 6046–6062 (2016).
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

