Rapid Evolution of Reduced Susceptibility against a Balanced Dual-Targeting Antibiotic through Stepping-Stone Mutations
- PMID: 31235632
- PMCID: PMC6709476
- DOI: 10.1128/AAC.00207-19
Rapid Evolution of Reduced Susceptibility against a Balanced Dual-Targeting Antibiotic through Stepping-Stone Mutations
Abstract
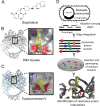
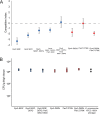
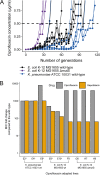
Multitargeting antibiotics, i.e., single compounds capable of inhibiting two or more bacterial targets, are generally considered to be a promising therapeutic strategy against resistance evolution. The rationale for this theory is that multitargeting antibiotics demand the simultaneous acquisition of multiple mutations at their respective target genes to achieve significant resistance. The theory presumes that individual mutations provide little or no benefit to the bacterial host. Here, we propose that such individual stepping-stone mutations can be prevalent in clinical bacterial isolates, as they provide significant resistance to other antimicrobial agents. To test this possibility, we focused on gepotidacin, an antibiotic candidate that selectively inhibits both bacterial DNA gyrase and topoisomerase IV. In a susceptible organism, Klebsiella pneumoniae, a combination of two specific mutations in these target proteins provide an >2,000-fold reduction in susceptibility, while individually, none of these mutations affect resistance significantly. Alarmingly, strains with decreased susceptibility against gepotidacin are found to be as virulent as the wild-type Klebsiella pneumoniae strain in a murine model. Moreover, numerous pathogenic isolates carry mutations which could promote the evolution of clinically significant reduction of susceptibility against gepotidacin in the future. As might be expected, prolonged exposure to ciprofloxacin, a clinically widely employed gyrase inhibitor, coselected for reduced susceptibility against gepotidacin. We conclude that extensive antibiotic usage could select for mutations that serve as stepping-stones toward resistance against antimicrobial compounds still under development. Our research indicates that even balanced multitargeting antibiotics are prone to resistance evolution.
Keywords: antibiotic resistance; genome engineering; gepotidacin.
Copyright © 2019 American Society for Microbiology.
Figures
Similar articles
-
Mechanistic and Structural Basis for the Actions of the Antibacterial Gepotidacin against Staphylococcus aureus Gyrase.ACS Infect Dis. 2019 Apr 12;5(4):570-581. doi: 10.1021/acsinfecdis.8b00315. Epub 2019 Feb 28. ACS Infect Dis. 2019. PMID: 30757898 Free PMC article.
-
Diverse phenotypic and genotypic characterization among clinical Klebsiella pneumoniae and Escherichia coli isolates carrying plasmid-mediated quinolone resistance determinants.Microb Drug Resist. 2011 Sep;17(3):363-7. doi: 10.1089/mdr.2011.0034. Epub 2011 May 12. Microb Drug Resist. 2011. PMID: 21563956
-
Frequency of DNA gyrase and topoisomerase IV mutations and plasmid-mediated quinolone resistance genes among Escherichia coli and Klebsiella pneumoniae isolated from urinary tract infections in Azerbaijan, Iran.J Glob Antimicrob Resist. 2019 Jun;17:39-43. doi: 10.1016/j.jgar.2018.11.003. Epub 2018 Nov 13. J Glob Antimicrob Resist. 2019. PMID: 30445211
-
Management of Neisseria gonorrhoeae infection: from drug resistance to drug repurposing.Expert Opin Ther Pat. 2024 Jun;34(6):511-524. doi: 10.1080/13543776.2024.2367005. Epub 2024 Jun 14. Expert Opin Ther Pat. 2024. PMID: 38856987 Review.
-
Dose Selection for Phase III Clinical Evaluation of Gepotidacin (GSK2140944) in the Treatment of Uncomplicated Urinary Tract Infections.Antimicrob Agents Chemother. 2022 Mar 15;66(3):e0149221. doi: 10.1128/AAC.01492-21. Epub 2022 Jan 3. Antimicrob Agents Chemother. 2022. PMID: 34978887 Free PMC article. Review.
Cited by
-
In silico gepotidacin target mining among 33 213 global Neisseria gonorrhoeae genomes from 1928 to 2023 combined with gepotidacin MIC testing of 22 gonococcal isolates with different GyrA and ParC substitutions.J Antimicrob Chemother. 2024 Sep 3;79(9):2221-2226. doi: 10.1093/jac/dkae217. J Antimicrob Chemother. 2024. PMID: 39004438 Free PMC article.
-
Recombineering and MAGE.Nat Rev Methods Primers. 2021;1:7. doi: 10.1038/s43586-020-00006-x. Epub 2021 Jan 14. Nat Rev Methods Primers. 2021. PMID: 35540496 Free PMC article.
-
Improved bacterial recombineering by parallelized protein discovery.Proc Natl Acad Sci U S A. 2020 Jun 16;117(24):13689-13698. doi: 10.1073/pnas.2001588117. Epub 2020 May 28. Proc Natl Acad Sci U S A. 2020. PMID: 32467157 Free PMC article.
-
Cryptic phenotypic variation emerges rapidly during the adaptive evolution of a carbapenemase.Nat Ecol Evol. 2025 Jul 30. doi: 10.1038/s41559-025-02804-6. Online ahead of print. Nat Ecol Evol. 2025. PMID: 40739338
-
Gyrase and Topoisomerase IV: Recycling Old Targets for New Antibacterials to Combat Fluoroquinolone Resistance.ACS Infect Dis. 2024 Apr 12;10(4):1097-1115. doi: 10.1021/acsinfecdis.4c00128. Epub 2024 Apr 2. ACS Infect Dis. 2024. PMID: 38564341 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

