Sall4 regulates neuromesodermal progenitors and their descendants during body elongation in mouse embryos
- PMID: 31235634
- PMCID: PMC6679359
- DOI: 10.1242/dev.177659
Sall4 regulates neuromesodermal progenitors and their descendants during body elongation in mouse embryos
Abstract
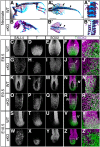
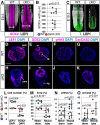
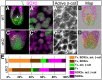
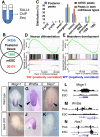
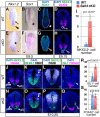
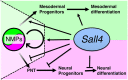
Bi-potential neuromesodermal progenitors (NMPs) produce both neural and paraxial mesodermal progenitors in the trunk and tail during vertebrate body elongation. We show that Sall4, a pluripotency-related transcription factor gene, has multiple roles in regulating NMPs and their descendants in post-gastrulation mouse embryos. Sall4 deletion using TCre caused body/tail truncation, reminiscent of early depletion of NMPs, suggesting a role of Sall4 in NMP maintenance. This phenotype became significant at the time of the trunk-to-tail transition, suggesting that Sall4 maintenance of NMPs enables tail formation. Sall4 mutants exhibit expanded neural and reduced mesodermal tissues, indicating a role of Sall4 in NMP differentiation balance. Mechanistically, we show that Sall4 promotion of WNT/β-catenin signaling contributes to NMP maintenance and differentiation balance. RNA-Seq and SALL4 ChIP-Seq analyses support the notion that Sall4 regulates both mesodermal and neural development. Furthermore, in the mesodermal compartment, genes regulating presomitic mesoderm differentiation are downregulated in Sall4 mutants. In the neural compartment, we show that differentiation of NMPs towards post-mitotic neuron is accelerated in Sall4 mutants. Our results collectively provide evidence supporting the role of Sall4 in regulating NMPs and their descendants.
Keywords: Body/tail elongation; Mesodermal progenitors and neural progenitors; Neuromesodermal progenitors; Sall4; WNT/β-catenin signaling.
© 2019. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

Similar articles
-
The Origin and Regulation of Neuromesodermal Progenitors (NMPs) in Embryos.Cells. 2024 Mar 21;13(6):549. doi: 10.3390/cells13060549. Cells. 2024. PMID: 38534393 Free PMC article. Review.
-
Lineage tracing of neuromesodermal progenitors reveals novel Wnt-dependent roles in trunk progenitor cell maintenance and differentiation.Development. 2015 May 1;142(9):1628-38. doi: 10.1242/dev.111922. Development. 2015. PMID: 25922526 Free PMC article.
-
Neuromesodermal specification during head-to-tail body axis formation.Curr Top Dev Biol. 2024;159:232-271. doi: 10.1016/bs.ctdb.2024.02.012. Epub 2024 Mar 19. Curr Top Dev Biol. 2024. PMID: 38729677 Review.
-
Dynamics of primitive streak regression controls the fate of neuromesodermal progenitors in the chicken embryo.Elife. 2021 Jul 6;10:e64819. doi: 10.7554/eLife.64819. Elife. 2021. PMID: 34227938 Free PMC article.
-
Sall4 regulates posterior trunk mesoderm development by promoting mesodermal gene expression and repressing neural genes in the mesoderm.Development. 2024 Mar 1;151(5):dev202649. doi: 10.1242/dev.202649. Epub 2024 Mar 4. Development. 2024. PMID: 38345319 Free PMC article.
Cited by
-
The Origin and Regulation of Neuromesodermal Progenitors (NMPs) in Embryos.Cells. 2024 Mar 21;13(6):549. doi: 10.3390/cells13060549. Cells. 2024. PMID: 38534393 Free PMC article. Review.
-
Regulation of Oct4 in stem cells and neural crest cells.Birth Defects Res. 2022 Oct 1;114(16):983-1002. doi: 10.1002/bdr2.2007. Epub 2022 Apr 1. Birth Defects Res. 2022. PMID: 35365980 Free PMC article. Review.
-
Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment.Nat Genet. 2022 Aug;54(8):1178-1191. doi: 10.1038/s41588-022-01134-8. Epub 2022 Jul 28. Nat Genet. 2022. PMID: 35902743 Free PMC article.
-
Sall4 restricts glycolytic metabolism in limb buds through transcriptional regulation of glycolytic enzyme genes.Dev Biol. 2023 Sep;501:28-38. doi: 10.1016/j.ydbio.2023.06.004. Epub 2023 Jun 8. Dev Biol. 2023. PMID: 37301463 Free PMC article.
-
METTL14 regulates chondrogenesis through the GDF5-RUNX-extracellular matrix gene axis during limb development.Nat Commun. 2025 Apr 30;16(1):4072. doi: 10.1038/s41467-025-59346-5. Nat Commun. 2025. PMID: 40307229 Free PMC article.
References
-
- Abu-Abed S., Dolle P., Metzger D., Beckett B., Chambon P. and Petkovich M. (2001). The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 15, 226-240. 10.1101/gad.855001 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

