Cryo-EM structures of four polymorphic TDP-43 amyloid cores
- PMID: 31235914
- PMCID: PMC7047951
- DOI: 10.1038/s41594-019-0248-4
Cryo-EM structures of four polymorphic TDP-43 amyloid cores
Abstract
The DNA and RNA processing protein TDP-43 undergoes both functional and pathogenic aggregation. Functional TDP-43 aggregates form reversible, transient species such as nuclear bodies, stress granules, and myo-granules. Pathogenic, irreversible TDP-43 aggregates form in amyotrophic lateral sclerosis and other neurodegenerative conditions. Here we find the features of TDP-43 fibrils that confer both reversibility and irreversibility by determining structures of two segments reported to be the pathogenic cores of human TDP-43 aggregation: SegA (residues 311-360), which forms three polymorphs, all with dagger-shaped folds; and SegB A315E (residues 286-331 containing the amyotrophic lateral sclerosis hereditary mutation A315E), which forms R-shaped folds. Energetic analysis suggests that the dagger-shaped polymorphs represent irreversible fibril structures, whereas the SegB polymorph may participate in both reversible and irreversible fibrils. Our structures reveal the polymorphic nature of TDP-43 and suggest how the A315E mutation converts the R-shaped polymorph to an irreversible form that enhances pathology.
Figures


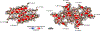
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

