Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism
- PMID: 31235964
- PMCID: PMC7368972
- DOI: 10.1038/s41591-019-0485-4
Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism
Abstract
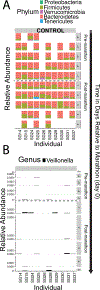
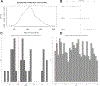
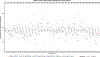
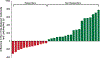
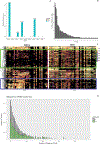
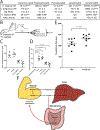
The human gut microbiome is linked to many states of human health and disease1. The metabolic repertoire of the gut microbiome is vast, but the health implications of these bacterial pathways are poorly understood. In this study, we identify a link between members of the genus Veillonella and exercise performance. We observed an increase in Veillonella relative abundance in marathon runners postmarathon and isolated a strain of Veillonella atypica from stool samples. Inoculation of this strain into mice significantly increased exhaustive treadmill run time. Veillonella utilize lactate as their sole carbon source, which prompted us to perform a shotgun metagenomic analysis in a cohort of elite athletes, finding that every gene in a major pathway metabolizing lactate to propionate is at higher relative abundance postexercise. Using 13C3-labeled lactate in mice, we demonstrate that serum lactate crosses the epithelial barrier into the lumen of the gut. We also show that intrarectal instillation of propionate is sufficient to reproduce the increased treadmill run time performance observed with V. atypica gavage. Taken together, these studies reveal that V. atypica improves run time via its metabolic conversion of exercise-induced lactate into propionate, thereby identifying a natural, microbiome-encoded enzymatic process that enhances athletic performance.
Conflict of interest statement
Competing Interests Statement
J.S. and G.M.C. are co-founders of FitBiomics, Inc. They and A.D.K. hold equity in FitBiomics, Inc.
Figures
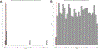

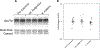

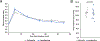

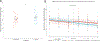

Comment in
-
Can microbes increase exercise performance in athletes?Nat Rev Endocrinol. 2019 Nov;15(11):629-630. doi: 10.1038/s41574-019-0250-2. Nat Rev Endocrinol. 2019. PMID: 31391569 No abstract available.
References
-
- Rothschild D et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). - PubMed
-
- Dusko Ehrlich S & The MetaHIT Consortium. MetaHIT: The European Union Project on Metagenomics of the Human Intestinal Tract in Metagenomics of the Human Body 307–316 (Springer, New York, NY, 2011). doi:10.1007/978-1-4419-7089-3_15 - DOI
Methods-Only References
-
- Team, R. C. & Others. R: A language and environment for statistical computing. (2013).
-
- Wickham H & Chang W ggplot2: An Implementation of the Grammar of Graphics. 2015. URL http://CRAN.R-project.org/package=ggplot2. R package version 1, (2015).
-
- Wickham H & Francois R dplyr: A grammar of data manipulation. R package version 0. 4 3, (2015).
-
- Murrell P The grid graphics package. R News 2, 14–19 (2002).
-
- Wickham H reshape2: Flexibly reshape data: a reboot of the reshape package. R package version. 2012; 1.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

