Pancreatic Ductal Organoids React Kras Dependent to the Removal of Tumor Suppressive Roadblocks
- PMID: 31236113
- PMCID: PMC6545725
- DOI: 10.1155/2019/2079742
Pancreatic Ductal Organoids React Kras Dependent to the Removal of Tumor Suppressive Roadblocks
Abstract
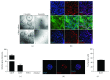
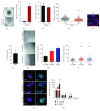
Pancreatic ductal adenocarcinoma (PDAC) is still the Achilles heel in modern oncology, with an increasing incidence accompanied by a persisting high mortality. The developmental process of PDAC is thought to be stepwise via precursor lesions and sequential accumulation of mutations. Thereby, current sequencing studies recapitulate this genetic heterogeneity in PDAC and show besides a handful of driver mutations (KRAS, TP53) a plethora of passenger mutations that allow to define subtypes. However, modeling the mutations of interest and their effects is still challenging. Interestingly, organoids have the potential to recapitulate in vitro, the in vivo characteristics of the tissue they originate from. Here, we could establish and develop tools allowing us to isolate, culture, and genetically modify ductal mouse organoids. Transferred to known effectors in the IPMN-PDAC sequence, we could reveal significantly increased proliferative and self-renewal capacities for PTEN and RNF43 deficiency in the context of oncogenic KRASG12D in mouse pancreatic organoids. Overall, we were able to obtain promising data centering ductal organoids in the focus of future PDAC research.
Figures

References
-
- Rahib L., Smith B. D., Aizenberg R., Rosenzweig A. B., Fleshman J. M., Matrisian L. M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Research. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. - DOI - PubMed
-
- Roberts N. J., Norris A. L., Petersen G. M., et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discovery. 2016;6(2):166–175. doi: 10.1158/2159-8290.CD-15-0402. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

