Immune and environment-driven gene expression during invasion: An eco-immunological application of RNA-Seq
- PMID: 31236254
- PMCID: PMC6580278
- DOI: 10.1002/ece3.5249
Immune and environment-driven gene expression during invasion: An eco-immunological application of RNA-Seq
Abstract
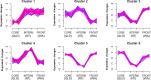
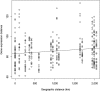
Host-pathogen associations change rapidly during a biological invasion and are predicted to impose strong selection on immune function. It has been proposed that the invader may experience an abrupt reduction in pathogen-mediated selection ("enemy release"), thereby favoring decreased investment into "costly" immune responses. Across plants and animals, there is mixed support for this prediction. Pathogens are not the only form of selection imposed on invaders; differences in abiotic environmental conditions between native and introduced ranges are also expected to drive rapid evolution. Here, we use RNA-Seq to assess the expression patterns of immune and environmentally associated genes in the cane toad (Rhinella marina) across its invasive Australian range. Transcripts encoding mediators of costly immune responses (inflammation, cytotoxicity) showed a curvilinear relationship with invasion history, with highest expression in toads from oldest and newest colonized areas. This pattern is surprising given theoretical expectations of density dynamics in invasive species and may be because density influences both intraspecific competition and parasite transmission, generating conflicting effects on the strength of immune responses. Alternatively, this expression pattern may be the result of other evolutionary forces, such as spatial sorting and genetic drift, working simultaneously with natural selection. Our findings do not support predictions about immune function based on the enemy release hypothesis and suggest instead that the effects of enemy release are difficult to isolate in wild populations, especially in the absence of information regarding parasite and pathogen infection. Additionally, expression patterns of genes underlying putatively environmentally associated traits are consistent with previous genetic studies, providing further support that Australian cane toads have adapted to novel abiotic challenges.
Keywords: Bufo marinus; cane toad; compositional data analysis; enemy release hypothesis; invasive species; spatial sorting.
Conflict of interest statement
None declared.
Figures


References
-
- Aitchison, J. , & Egozcue, J. J. (2005). Compositional data analysis: Where are we and where should we be heading? Mathematical Geology, 37, 829–850. 10.1007/s11004-005-7383-7 - DOI
-
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Retrieved from http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Bax, N. , Williamson, A. , Aguero, M. , Gonzalez, E. , & Geeves, W. (2003). Marine invasive alien species: A threat to global biodiversity. Marine Policy, 27, 313–323. 10.1016/S0308-597X(03)00041-1 - DOI
LinkOut - more resources
Full Text Sources

