Intertumoral heterogeneity in patient-specific drug sensitivities in treatment-naïve glioblastoma
- PMID: 31238897
- PMCID: PMC6593575
- DOI: 10.1186/s12885-019-5861-4
Intertumoral heterogeneity in patient-specific drug sensitivities in treatment-naïve glioblastoma
Abstract
Background: A major barrier to effective treatment of glioblastoma (GBM) is the large intertumoral heterogeneity at the genetic and cellular level. In early phase clinical trials, patient heterogeneity in response to therapy is commonly observed; however, how tumor heterogeneity is reflected in individual drug sensitivities in the treatment-naïve glioblastoma stem cells (GSC) is unclear.
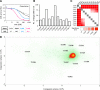
Methods: We cultured 12 patient-derived primary GBMs as tumorspheres and validated tumor stem cell properties by functional assays. Using automated high-throughput screening (HTS), we evaluated sensitivity to 461 anticancer drugs in a collection covering most FDA-approved anticancer drugs and investigational compounds with a broad range of molecular targets. Statistical analyses were performed using one-way ANOVA and Spearman correlation.
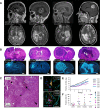
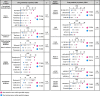
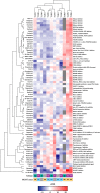
Results: Although tumor stem cell properties were confirmed in GSC cultures, their in vitro and in vivo morphology and behavior displayed considerable tumor-to-tumor variability. Drug screening revealed significant differences in the sensitivity to anticancer drugs (p < 0.0001). The patient-specific vulnerabilities to anticancer drugs displayed a heterogeneous pattern. They represented a variety of mechanistic drug classes, including apoptotic modulators, conventional chemotherapies, and inhibitors of histone deacetylases, heat shock proteins, proteasomes and different kinases. However, the individual GSC cultures displayed high biological consistency in drug sensitivity patterns within a class of drugs. An independent laboratory confirmed individual drug responses.
Conclusions: This study demonstrates that patient-derived and treatment-naïve GSC cultures maintain patient-specific traits and display intertumoral heterogeneity in drug sensitivity to anticancer drugs. The heterogeneity in patient-specific drug responses highlights the difficulty in applying targeted treatment strategies at the population level to GBM patients. However, HTS can be applied to uncover patient-specific drug sensitivities for functional precision medicine.
Keywords: Drug sensitivity; Functional precision medicine; Glioblastoma; Glioblastoma stem cells; High-throughput drug screening; Individualized medicine.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Feasibility study of using high-throughput drug sensitivity testing to target recurrent glioblastoma stem cells for individualized treatment.Clin Transl Med. 2019 Dec 30;8(1):33. doi: 10.1186/s40169-019-0253-6. Clin Transl Med. 2019. PMID: 31889236 Free PMC article.
-
The efficacy of a coordinated pharmacological blockade in glioblastoma stem cells with nine repurposed drugs using the CUSP9 strategy.J Cancer Res Clin Oncol. 2019 Jun;145(6):1495-1507. doi: 10.1007/s00432-019-02920-4. Epub 2019 Apr 26. J Cancer Res Clin Oncol. 2019. PMID: 31028540 Free PMC article.
-
Molecular heterogeneity in a patient-derived glioblastoma xenoline is regulated by different cancer stem cell populations.PLoS One. 2015 May 8;10(5):e0125838. doi: 10.1371/journal.pone.0125838. eCollection 2015. PLoS One. 2015. PMID: 25955030 Free PMC article.
-
Rapid identification and validation of novel targeted approaches for Glioblastoma: A combined ex vivo-in vivo pharmaco-omic model.Exp Neurol. 2018 Jan;299(Pt B):281-288. doi: 10.1016/j.expneurol.2017.09.006. Epub 2017 Sep 18. Exp Neurol. 2018. PMID: 28923369 Review.
-
Advances in histone deacetylase inhibitors in targeting glioblastoma stem cells.Cancer Chemother Pharmacol. 2020 Aug;86(2):165-179. doi: 10.1007/s00280-020-04109-w. Epub 2020 Jul 7. Cancer Chemother Pharmacol. 2020. PMID: 32638092 Review.
Cited by
-
Leveraging multi-omics data to infer regulators of mRNA 3' end processing in glioblastoma.Front Mol Biosci. 2024 Aug 12;11:1363933. doi: 10.3389/fmolb.2024.1363933. eCollection 2024. Front Mol Biosci. 2024. PMID: 39188787 Free PMC article.
-
Involvement of Phosphatase and Tensin Homolog in Cyclin-Dependent Kinase 4/6 Inhibitor-Induced Blockade of Glioblastoma.Front Pharmacol. 2019 Nov 7;10:1316. doi: 10.3389/fphar.2019.01316. eCollection 2019. Front Pharmacol. 2019. PMID: 31787897 Free PMC article.
-
Association of plasma cell-free DNA with survival in patients with IDH wild-type glioblastoma.Neurooncol Adv. 2021 Jan 16;3(1):vdab011. doi: 10.1093/noajnl/vdab011. eCollection 2021 Jan-Dec. Neurooncol Adv. 2021. PMID: 33615225 Free PMC article.
-
Functional Precision Oncology: The Next Frontier to Improve Glioblastoma Outcome?Int J Mol Sci. 2022 Aug 3;23(15):8637. doi: 10.3390/ijms23158637. Int J Mol Sci. 2022. PMID: 35955765 Free PMC article. Review.
-
p53 Signaling on Microenvironment and Its Contribution to Tissue Chemoresistance.Membranes (Basel). 2022 Feb 9;12(2):202. doi: 10.3390/membranes12020202. Membranes (Basel). 2022. PMID: 35207121 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

