Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli
- PMID: 31239340
- PMCID: PMC6628675
- DOI: 10.1073/pnas.1904428116
Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli
Erratum in
-
Correction for Hu et al., Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli.Proc Natl Acad Sci U S A. 2020 Jan 21;117(3):1819. doi: 10.1073/pnas.1922471117. Epub 2020 Jan 13. Proc Natl Acad Sci U S A. 2020. PMID: 31932431 Free PMC article. No abstract available.
Abstract
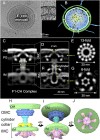
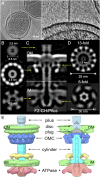
Bacterial conjugation systems are members of the large type IV secretion system (T4SS) superfamily. Conjugative transfer of F plasmids residing in the Enterobacteriaceae was first reported in the 1940s, yet the architecture of F plasmid-encoded transfer channel and its physical relationship with the F pilus remain unknown. We visualized F-encoded structures in the native bacterial cell envelope by in situ cryoelectron tomography (CryoET). Remarkably, F plasmids encode four distinct structures, not just the translocation channel or channel-pilus complex predicted by prevailing models. The F1 structure is composed of distinct outer and inner membrane complexes and a connecting cylinder that together house the envelope-spanning translocation channel. The F2 structure is essentially the F1 complex with the F pilus attached at the outer membrane (OM). Remarkably, the F3 structure consists of the F pilus attached to a thin, cell envelope-spanning stalk, whereas the F4 structure consists of the pilus docked to the OM without an associated periplasmic density. The traffic ATPase TraC is configured as a hexamer of dimers at the cytoplasmic faces of the F1 and F2 structures, where it respectively regulates substrate transfer and F pilus biogenesis. Together, our findings present architectural renderings of the DNA conjugation or "mating" channel, the channel-pilus connection, and unprecedented pilus basal structures. These structural snapshots support a model for biogenesis of the F transfer system and allow for detailed comparisons with other structurally characterized T4SSs.
Keywords: DNA conjugation; cryoelectron tomography; pilus; protein transport; type IV secretion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
In Situ Visualization of the pKM101-Encoded Type IV Secretion System Reveals a Highly Symmetric ATPase Energy Center.mBio. 2021 Oct 26;12(5):e0246521. doi: 10.1128/mBio.02465-21. Epub 2021 Oct 12. mBio. 2021. PMID: 34634937 Free PMC article.
-
Cryo-EM Structure of the relaxosome, a complex essential for bacterial mating and the spread of antibiotic resistance genes.Nat Commun. 2025 May 27;16(1):4906. doi: 10.1038/s41467-025-60116-6. Nat Commun. 2025. PMID: 40425557 Free PMC article.
-
Contributions of F-specific subunits to the F plasmid-encoded type IV secretion system and F pilus.Mol Microbiol. 2022 May;117(5):1275-1290. doi: 10.1111/mmi.14908. Epub 2022 Apr 26. Mol Microbiol. 2022. PMID: 35434837 Free PMC article.
-
F conjugation: back to the beginning.Plasmid. 2013 Jul;70(1):18-32. doi: 10.1016/j.plasmid.2013.03.010. Epub 2013 Apr 28. Plasmid. 2013. PMID: 23632276 Review.
-
Analysis of the sequence and gene products of the transfer region of the F sex factor.Microbiol Rev. 1994 Jun;58(2):162-210. doi: 10.1128/mr.58.2.162-210.1994. Microbiol Rev. 1994. PMID: 7915817 Free PMC article. Review.
Cited by
-
Subinhibitory antibiotic concentrations promote the horizontal transfer of plasmid-borne resistance genes from Klebsiellae pneumoniae to Escherichia coli.Front Microbiol. 2022 Nov 7;13:1017092. doi: 10.3389/fmicb.2022.1017092. eCollection 2022. Front Microbiol. 2022. PMID: 36419429 Free PMC article.
-
Real-time visualisation of the intracellular dynamics of conjugative plasmid transfer.Nat Commun. 2023 Jan 18;14(1):294. doi: 10.1038/s41467-023-35978-3. Nat Commun. 2023. PMID: 36653393 Free PMC article.
-
Morphological remodeling of Coxiella burnetii during its biphasic developmental cycle revealed by cryo-electron tomography.iScience. 2023 Jun 24;26(7):107210. doi: 10.1016/j.isci.2023.107210. eCollection 2023 Jul 21. iScience. 2023. PMID: 37485371 Free PMC article.
-
The Helicobacter pylori Cag Type IV Secretion System.Trends Microbiol. 2020 Aug;28(8):682-695. doi: 10.1016/j.tim.2020.02.004. Epub 2020 Mar 26. Trends Microbiol. 2020. PMID: 32451226 Free PMC article. Review.
-
Coexistence of a nonresistance-conferring IncI1 plasmid favors persistence of the blaCTX-M-bearing IncFII plasmid in Escherichia coli.Microbiol Spectr. 2024 Jun 4;12(6):e0424023. doi: 10.1128/spectrum.04240-23. Epub 2024 Apr 30. Microbiol Spectr. 2024. PMID: 38687059 Free PMC article.
References
-
- Lederberg J., Tatum E. L., Gene recombination in Escherichia coli. Nature 158, 558 (1946). - PubMed
-
- Cabezón E., Ripoll-Rozada J., Peña A., de la Cruz F., Arechaga I., Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 39, 81–95 (2015). - PubMed
-
- Frost L. S., Koraimann G., Regulation of bacterial conjugation: Balancing opportunity with adversity. Future Microbiol. 5, 1057–1071 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

