A Lotus japonicus cytoplasmic kinase connects Nod factor perception by the NFR5 LysM receptor to nodulation
- PMID: 31239345
- PMCID: PMC6628658
- DOI: 10.1073/pnas.1815425116
A Lotus japonicus cytoplasmic kinase connects Nod factor perception by the NFR5 LysM receptor to nodulation
Abstract
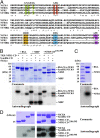
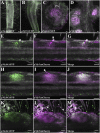
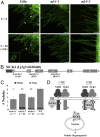
The establishment of nitrogen-fixing root nodules in legume-rhizobia symbiosis requires an intricate communication between the host plant and its symbiont. We are, however, limited in our understanding of the symbiosis signaling process. In particular, how membrane-localized receptors of legumes activate signal transduction following perception of rhizobial signaling molecules has mostly remained elusive. To address this, we performed a coimmunoprecipitation-based proteomics screen to identify proteins associated with Nod factor receptor 5 (NFR5) in Lotus japonicus. Out of 51 NFR5-associated proteins, we focused on a receptor-like cytoplasmic kinase (RLCK), which we named NFR5-interacting cytoplasmic kinase 4 (NiCK4). NiCK4 associates with heterologously expressed NFR5 in Nicotiana benthamiana, and directly binds and phosphorylates the cytoplasmic domains of NFR5 and NFR1 in vitro. At the cellular level, Nick4 is coexpressed with Nfr5 in root hairs and nodule cells, and the NiCK4 protein relocates to the nucleus in an NFR5/NFR1-dependent manner upon Nod factor treatment. Phenotyping of retrotransposon insertion mutants revealed that NiCK4 promotes nodule organogenesis. Together, these results suggest that the identified RLCK, NiCK4, acts as a component of the Nod factor signaling pathway downstream of NFR5.
Keywords: Lotus; NFR5; NiCK4; RLCK; nodulation.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases.Elife. 2014 Nov 25;3:e03891. doi: 10.7554/eLife.03891. Elife. 2014. PMID: 25422918 Free PMC article.
-
LACK OF SYMBIONT ACCOMMODATION controls intracellular symbiont accommodation in root nodule and arbuscular mycorrhizal symbiosis in Lotus japonicus.PLoS Genet. 2019 Jan 3;15(1):e1007865. doi: 10.1371/journal.pgen.1007865. eCollection 2019 Jan. PLoS Genet. 2019. PMID: 30605473 Free PMC article.
-
Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus.Elife. 2018 Jun 29;7:e33506. doi: 10.7554/eLife.33506. Elife. 2018. PMID: 29957177 Free PMC article.
-
How many peas in a pod? Legume genes responsible for mutualistic symbioses underground.Plant Cell Physiol. 2010 Sep;51(9):1381-97. doi: 10.1093/pcp/pcq107. Epub 2010 Jul 21. Plant Cell Physiol. 2010. PMID: 20660226 Free PMC article. Review.
-
Evolution of NIN and NIN-like Genes in Relation to Nodule Symbiosis.Genes (Basel). 2020 Jul 11;11(7):777. doi: 10.3390/genes11070777. Genes (Basel). 2020. PMID: 32664480 Free PMC article. Review.
Cited by
-
Signaling in Legume-Rhizobia Symbiosis.Int J Mol Sci. 2023 Dec 12;24(24):17397. doi: 10.3390/ijms242417397. Int J Mol Sci. 2023. PMID: 38139226 Free PMC article. Review.
-
Innovation and appropriation in mycorrhizal and rhizobial Symbioses.Plant Cell. 2022 Apr 26;34(5):1573-1599. doi: 10.1093/plcell/koac039. Plant Cell. 2022. PMID: 35157080 Free PMC article.
-
Role of Nod factor receptors and its allies involved in nitrogen fixation.Planta. 2023 Feb 13;257(3):54. doi: 10.1007/s00425-023-04090-7. Planta. 2023. PMID: 36780015 Review.
-
Development versus predation: Transcriptomic changes during the lifecycle of Myxococcus xanthus.Front Microbiol. 2022 Sep 26;13:1004476. doi: 10.3389/fmicb.2022.1004476. eCollection 2022. Front Microbiol. 2022. PMID: 36225384 Free PMC article.
-
Establishment of Proximity-Dependent Biotinylation Approaches in Different Plant Model Systems.Plant Cell. 2020 Nov;32(11):3388-3407. doi: 10.1105/tpc.20.00235. Epub 2020 Aug 25. Plant Cell. 2020. PMID: 32843435 Free PMC article.
References
-
- Fisher R. F., Long S. R., Rhizobium–plant signal exchange. Nature 357, 655–660 (1992). - PubMed
-
- Denarie J., et al. , “Rhizobium and legume nodulation—a molecular dialog” in New Horizons in Nitrogen Fixation. Current Plant Science and Biotechnology in Agriculture, R. Palacios, J. Mora, W. E. Newton, Eds. (Springer, Dordrecht, 1993), vol. 17, pp. 19–30.
-
- Madsen E. B., et al. , A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

