Ribosome Biogenesis in Plants: From Functional 45S Ribosomal DNA Organization to Ribosome Assembly Factors
- PMID: 31239391
- PMCID: PMC6751116
- DOI: 10.1105/tpc.18.00874
Ribosome Biogenesis in Plants: From Functional 45S Ribosomal DNA Organization to Ribosome Assembly Factors
Abstract
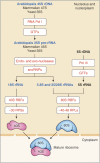
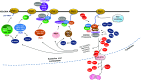
The transcription of 18S, 5.8S, and 18S rRNA genes (45S rDNA), cotranscriptional processing of pre-rRNA, and assembly of mature rRNA with ribosomal proteins are the linchpins of ribosome biogenesis. In yeast (Saccharomyces cerevisiae) and animal cells, hundreds of pre-rRNA processing factors have been identified and their involvement in ribosome assembly determined. These studies, together with structural analyses, have yielded comprehensive models of the pre-40S and pre-60S ribosome subunits as well as the largest cotranscriptionally assembled preribosome particle: the 90S/small subunit processome. Here, we present the current knowledge of the functional organization of 45S rDNA, pre-rRNA transcription, rRNA processing activities, and ribosome assembly factors in plants, focusing on data from Arabidopsis (Arabidopsis thaliana). Based on yeast and mammalian cell studies, we describe the ribonucleoprotein complexes and RNA-associated activities and discuss how they might specifically affect the production of 40S and 60S subunits. Finally, we review recent findings concerning pre-rRNA processing pathways and a novel mechanism involved in a ribosome stress response in plants.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures



References
-
- Abbasi N., Kim H.B., Park N.I., Kim H.S., Kim Y.K., Park Y.I., Choi S.B. (2010). APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 64: 960–976. - PubMed
-
- Abou-Ellail M., Cooke R., Sáez-Vásquez J. (2011). Variations in a team: Major and minor variants of Arabidopsis thaliana rDNA genes. Nucleus 2: 294–299. - PubMed
-
- Ahn C.S., Cho H.K., Lee D.H., Sim H.J., Kim S.G., Pai H.S. (2016). Functional characterization of the ribosome biogenesis factors PES, BOP1, and WDR12 (PeBoW), and mechanisms of defective cell growth and proliferation caused by PeBoW deficiency in Arabidopsis. J. Exp. Bot. 67: 5217–5232. - PMC - PubMed
-
- Albert B., Colleran C., Léger-Silvestre I., Berger A.B., Dez C., Normand C., Perez-Fernandez J., McStay B., Gadal O. (2013). Structure-function analysis of Hmo1 unveils an ancestral organization of HMG-box factors involved in ribosomal DNA transcription from yeast to human. Nucleic Acids Res. 41: 10135–10149. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

