MDR: an integrative DNA N6-methyladenine and N4-methylcytosine modification database for Rosaceae
- PMID: 31240103
- PMCID: PMC6572862
- DOI: 10.1038/s41438-019-0160-4
MDR: an integrative DNA N6-methyladenine and N4-methylcytosine modification database for Rosaceae
Abstract
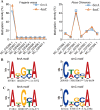
Eukaryotic DNA methylation has been receiving increasing attention for its crucial epigenetic regulatory function. The recently developed single-molecule real-time (SMRT) sequencing technology provides an efficient way to detect DNA N6-methyladenine (6mA) and N4-methylcytosine (4mC) modifications at a single-nucleotide resolution. The family Rosaceae contains horticultural plants with a wide range of economic importance. However, little is currently known regarding the genome-wide distribution patterns and functions of 6mA and 4mC modifications in the Rosaceae. In this study, we present an integrated DNA 6mA and 4mC modification database for the Rosaceae (MDR, http://mdr.xieslab.org). MDR, the first repository for displaying and storing DNA 6mA and 4mC methylomes from SMRT sequencing data sets for Rosaceae, includes meta and statistical information, methylation densities, Gene Ontology enrichment analyses, and genome search and browse for methylated sites in NCBI. MDR provides important information regarding DNA 6mA and 4mC methylation and may help users better understand epigenetic modifications in the family Rosaceae.
Keywords: DNA methylation; DNA sequencing; Plant genetics.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

