Paternal exposure to benzo(a)pyrene induces genome-wide mutations in mouse offspring
- PMID: 31240266
- PMCID: PMC6586636
- DOI: 10.1038/s42003-019-0476-5
Paternal exposure to benzo(a)pyrene induces genome-wide mutations in mouse offspring
Abstract
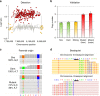
Understanding the effects of environmental exposures on germline mutation rates has been a decades-long pursuit in genetics. We used next-generation sequencing and comparative genomic hybridization arrays to investigate genome-wide mutations in the offspring of male mice exposed to benzo(a)pyrene (BaP), a common environmental pollutant. We demonstrate that offspring developing from sperm exposed during the mitotic or post-mitotic phases of spermatogenesis have significantly more de novo single nucleotide variants (1.8-fold; P < 0.01) than controls. Both phases of spermatogenesis are susceptible to the induction of heritable mutations, although mutations arising from post-fertilization events are more common after post-mitotic exposure. In addition, the mutation spectra in sperm and offspring of BaP-exposed males are consistent. Finally, we report a significant increase in transmitted copy number duplications (P = 0.001) in BaP-exposed sires. Our study demonstrates that germ cell mutagen exposures induce genome-wide mutations in the offspring that may be associated with adverse health outcomes.
Keywords: Genomics; Mutation.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

