NMDA Receptors Regulate Neuregulin 2 Binding to ER-PM Junctions and Ectodomain Release by ADAM10 [corrected]
- PMID: 31240601
- PMCID: PMC8262096
- DOI: 10.1007/s12035-019-01659-w
NMDA Receptors Regulate Neuregulin 2 Binding to ER-PM Junctions and Ectodomain Release by ADAM10 [corrected]
Erratum in
-
Correction to: NMDA Receptors Regulate Neuregulin 2 Binding to ER-PM Junctions and Ectodomain Release by ADAM10.Mol Neurobiol. 2020 Jan;57(1):585. doi: 10.1007/s12035-019-01778-4. Mol Neurobiol. 2020. PMID: 31522381
Abstract
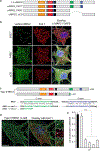
Unprocessed pro-neuregulin 2 (pro-NRG2) accumulates on neuronal cell bodies at junctions between the endoplasmic reticulum and plasma membrane (ER-PM junctions). NMDA receptors (NMDARs) trigger NRG2 ectodomain shedding from these sites followed by activation of ErbB4 receptor tyrosine kinases, and ErbB4 signaling cell-autonomously downregulates intrinsic excitability of GABAergic interneurons by reducing voltage-gated sodium channel currents. NMDARs also promote dispersal of Kv2.1 clusters from ER-PM junctions and cause a hyperpolarizing shift in its voltage-dependent channel activation, suggesting that NRG2/ErbB4 and Kv2.1 work together to regulate intrinsic interneuron excitability in an activity-dependent manner. Here we explored the cellular processes underlying NMDAR-dependent NRG2 shedding in cultured rat hippocampal neurons. We report that NMDARs control shedding by two separate but converging mechanisms. First, NMDA treatment disrupts binding of pro-NRG2 to ER-PM junctions by post-translationally modifying conserved Ser/Thr residues in its intracellular domain. Second, using a mutant NRG2 protein that cannot be modified at these residues and that fails to accumulate at ER-PM junctions, we demonstrate that NMDARs also directly promote NRG2 shedding by ADAM-type metalloproteinases. Using pharmacological and shRNA-mediated knockdown, and metalloproteinase overexpression, we unexpectedly find that ADAM10, but not ADAM17/TACE, is the major NRG2 sheddase acting downstream of NMDAR activation. Together, these findings reveal how NMDARs exert tight control over the NRG2/ErbB4 signaling pathway, and suggest that NRG2 and Kv2.1 are co-regulated components of a shared pathway that responds to elevated extracellular glutamate levels.
Keywords: ADAM10; Activity-dependent; ER-PM junction; Kv2.1; Neuregulin; Sheddase.
Figures
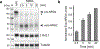

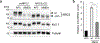



Similar articles
-
ER-PM Junctions on GABAergic Interneurons Are Organized by Neuregulin 2/VAP Interactions and Regulated by NMDA Receptors.Int J Mol Sci. 2023 Feb 2;24(3):2908. doi: 10.3390/ijms24032908. Int J Mol Sci. 2023. PMID: 36769244 Free PMC article.
-
Identification of VAPA and VAPB as Kv2 Channel-Interacting Proteins Defining Endoplasmic Reticulum-Plasma Membrane Junctions in Mammalian Brain Neurons.J Neurosci. 2018 Aug 29;38(35):7562-7584. doi: 10.1523/JNEUROSCI.0893-18.2018. Epub 2018 Jul 16. J Neurosci. 2018. PMID: 30012696 Free PMC article.
-
A negative feedback loop controls NMDA receptor function in cortical interneurons via neuregulin 2/ErbB4 signalling.Nat Commun. 2015 Jun 1;6:7222. doi: 10.1038/ncomms8222. Nat Commun. 2015. PMID: 26027736 Free PMC article.
-
Kv2 channels create endoplasmic reticulum / plasma membrane junctions: a brief history of Kv2 channel subcellular localization.Channels (Austin). 2019 Dec;13(1):88-101. doi: 10.1080/19336950.2019.1568824. Channels (Austin). 2019. PMID: 30712450 Free PMC article. Review.
-
Regulation and Role of Store-Operated Ca2+ Entry in Cellular Proliferation.In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 12. In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 12. PMID: 30299656 Free Books & Documents. Review.
Cited by
-
Neuregulins 1, 2, and 3 Promote Early Neurite Outgrowth in ErbB4-Expressing Cortical GABAergic Interneurons.Mol Neurobiol. 2020 Aug;57(8):3568-3588. doi: 10.1007/s12035-020-01966-7. Epub 2020 Jun 16. Mol Neurobiol. 2020. PMID: 32542595
-
Transcytosis and trans-synaptic retention by postsynaptic ErbB4 underlie axonal accumulation of NRG3.J Cell Biol. 2022 Jul 4;221(7):e202110167. doi: 10.1083/jcb.202110167. Epub 2022 May 17. J Cell Biol. 2022. PMID: 35579602 Free PMC article.
-
Proteolytic Processing of Neuregulin 2.Mol Neurobiol. 2020 Apr;57(4):1799-1813. doi: 10.1007/s12035-019-01846-9. Epub 2019 Dec 14. Mol Neurobiol. 2020. PMID: 31838721 Free PMC article.
-
ER-PM Junctions on GABAergic Interneurons Are Organized by Neuregulin 2/VAP Interactions and Regulated by NMDA Receptors.Int J Mol Sci. 2023 Feb 2;24(3):2908. doi: 10.3390/ijms24032908. Int J Mol Sci. 2023. PMID: 36769244 Free PMC article.
References
-
- Gerecke KM, Wyss JM, Karavanova I, Buonanno A, Carroll SL (2001) ErbB transmembrane tyrosine kinase receptors are differentially expressed throughout the adult rat central nervous system. J Comp Neurol 433 (1):86–100 - PubMed
-
- Buonanno A, Fischbach GD (2001) Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr Opin Neurobiol 11 (3):287–296. doi:S0959–4388(00)00210–5 [pii] - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

