What can be learned by scanning the genome for molecular convergence in wild populations?
- PMID: 31241191
- PMCID: PMC7586825
- DOI: 10.1111/nyas.14177
What can be learned by scanning the genome for molecular convergence in wild populations?
Abstract
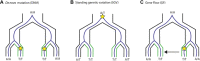
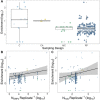
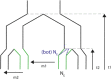
Convergent evolution, where independent lineages evolve similar phenotypes in response to similar challenges, can provide valuable insight into how selection operates and the limitations it encounters. However, it has only recently become possible to explore how convergent evolution is reflected at the genomic level. The overlapping outlier approach (OOA), where genome scans of multiple independent lineages are used to find outliers that overlap and therefore identify convergently evolving loci, is becoming popular. Here, we present a quantitative analysis of 34 studies that used this approach across many sampling designs, taxa, and sampling intensities. We found that OOA studies with increased biological sampling power within replicates have increased likelihood of finding overlapping, "convergent" signals of adaptation between them. When identifying convergent loci as overlapping outliers, it is tempting to assume that any false-positive outliers derived from individual scans will fail to overlap across replicates, but this cannot be guaranteed. We highlight how population demographics and genomic context can contribute toward both true convergence and false positives in OOA studies. We finish with an exploration of emerging methods that couple genome scans with phenotype and environmental measures, leveraging added information from genome data to more directly test hypotheses of the likelihood of convergent evolution.
Keywords: convergent evolution; genome scans; parallel evolution; population genomics.
© 2019 The Authors. Annals of the New York Academy of Sciences published by Wiley Periodicals, Inc. on behalf of New York Academy of Sciences.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Bell, M.A. & Foster S.A.. 1994. The Evolutionary Biology of the Threespine Stickleback. Oxford University Press.
-
- Heyduk, K. , Moreno‐Villena J.J., Gilman I.S., et al 2019. The genetics of convergent evolution: insights from plant photosynthesis. Nat. Rev. Genet. - PubMed
-
- Losos, J.B. 2011. Convergence, adaptation, and constraint. Evolution 65: 1827–1840. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

