Optimization of Replication, Transcription, and Translation in a Semi-Synthetic Organism
- PMID: 31241334
- PMCID: PMC6693872
- DOI: 10.1021/jacs.9b02075
Optimization of Replication, Transcription, and Translation in a Semi-Synthetic Organism
Abstract
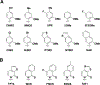
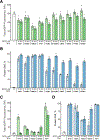
Previously, we reported the creation of a semi-synthetic organism (SSO) that stores and retrieves increased information by virtue of stably maintaining an unnatural base pair (UBP) in its DNA, transcribing the corresponding unnatural nucleotides into the codons and anticodons of mRNAs and tRNAs, and then using them to produce proteins containing noncanonical amino acids (ncAAs). Here we report a systematic extension of the effort to optimize the SSO by exploring a variety of deoxy- and ribonucleotide analogues. Importantly, this includes the first in vivo structure-activity relationship (SAR) analysis of unnatural ribonucleoside triphosphates. Similarities and differences between how DNA and RNA polymerases recognize the unnatural nucleotides were observed, and remarkably, we found that a wide variety of unnatural ribonucleotides can be efficiently transcribed into RNA and then productively and selectively paired at the ribosome to mediate the synthesis of proteins with ncAAs. The results extend previous studies, demonstrating that nucleotides bearing no significant structural or functional homology to the natural nucleotides can be efficiently and selectively paired during replication, to include each step of the entire process of information storage and retrieval. From a practical perspective, the results identify the most optimal UBP for replication and transcription, as well as the most optimal unnatural ribonucleoside triphosphates for transcription and translation. The optimized SSO is now, for the first time, able to efficiently produce proteins containing multiple, proximal ncAAs.
Conflict of interest statement
Notes
The authors declare the following competing financial interests: a patent application has been filed based on the use of UBPs in SSOs and F.E.R. has a financial interest (shares) in Synthorx, Inc., a company that has commercial interests in the UBP.
Figures





References
-
- Leduc S, The Mechanisms of Life Rebman Company: New York, 1911.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

