Inferring the distribution of fitness effects of spontaneous mutations in Chlamydomonas reinhardtii
- PMID: 31242179
- PMCID: PMC6615632
- DOI: 10.1371/journal.pbio.3000192
Inferring the distribution of fitness effects of spontaneous mutations in Chlamydomonas reinhardtii
Abstract
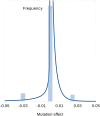
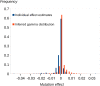
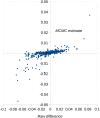
Spontaneous mutations are the source of new genetic variation and are thus central to the evolutionary process. In molecular evolution and quantitative genetics, the nature of genetic variation depends critically on the distribution of effects of mutations on fitness and other quantitative traits. Spontaneous mutation accumulation (MA) experiments have been the principal approach for investigating the overall rate of occurrence and cumulative effect of mutations but have not allowed the phenotypic effects of individual mutations to be studied directly. Here, we crossed MA lines of the green alga Chlamydomonas reinhardtii with its unmutated ancestral strain to create haploid recombinant lines, each carrying an average of 50% of the accumulated mutations in a large number of combinations. With the aid of the genome sequences of the MA lines, we inferred the genotypes of the mutations, assayed their growth rate as a measure of fitness, and inferred the distribution of fitness effects (DFE) using a Bayesian mixture model. We infer that the DFE is highly leptokurtic (L-shaped). Of mutations with absolute fitness effects exceeding 1%, about one-sixth increase fitness in the laboratory environment. The inferred distribution of effects for deleterious mutations is consistent with a strong role for nearly neutral evolution. Specifically, such a distribution predicts that nucleotide variation and genetic variation for quantitative traits will be insensitive to change in the effective population size.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Ohta T. Extension of the neutral mutation drift hypothesis In: Kimura M, editor. Molecular Evolution and Polymorphism. Mishima: National Institute of Genetics; 1977. pp. 148–167.
-
- Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press; 1983.
-
- Robertson A. The nature of quantitative genetic variation Heritage from Mendel. In: Brink RB, editor. Madison, Milwaukee, and London: University of Wisconsin Press; 1967. pp. 265–280.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

