The effect of breed and diet type on the global transcriptome of hepatic tissue in beef cattle divergent for feed efficiency
- PMID: 31242854
- PMCID: PMC6593537
- DOI: 10.1186/s12864-019-5906-8
The effect of breed and diet type on the global transcriptome of hepatic tissue in beef cattle divergent for feed efficiency
Abstract
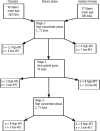
Background: Feed efficiency is an important economic and environmental trait in beef production, which can be measured in terms of residual feed intake (RFI). Cattle selected for low-RFI (feed efficient) have similar production levels but decreased feed intake, while also emitting less methane. RFI is difficult and expensive to measure and is not widely adopted in beef production systems. However, development of DNA-based biomarkers for RFI may facilitate its adoption in genomic-assisted breeding programmes. Cattle have been shown to re-rank in terms of RFI across diets and age, while also RFI varies by breed. Therefore, we used RNA-Seq technology to investigate the hepatic transcriptome of RFI-divergent Charolais (CH) and Holstein-Friesian (HF) steers across three dietary phases to identify genes and biological pathways associated with RFI regardless of diet or breed.
Results: Residual feed intake was measured during a high-concentrate phase, a zero-grazed grass phase and a final high-concentrate phase. In total, 322 and 33 differentially expressed genes (DEGs) were identified across all diets for CH and HF steers, respectively. Three genes, GADD45G, HP and MID1IP1, were differentially expressed in CH when both the high-concentrate zero-grazed grass diet were offered. Two canonical pathways were enriched across all diets for CH steers. These canonical pathways were related to immune function.
Conclusions: The absence of common differentially expressed genes across all dietary phases and breeds in this study supports previous reports of the re-ranking of animals in terms of RFI when offered differing diets over their lifetime. However, we have identified biological processes such as the immune response and lipid metabolism as potentially associated with RFI divergence emphasising the previously reported roles of these biological processes with respect to RFI.
Keywords: Bovine genetics; Feed efficiency; RNA-Seq.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
An across breed, diet and tissue analysis reveals the transcription factor NR1H3 as a key mediator of residual feed intake in beef cattle.BMC Genomics. 2024 Mar 4;25(1):234. doi: 10.1186/s12864-024-10151-2. BMC Genomics. 2024. PMID: 38438858 Free PMC article.
-
Effect of breed and diet on the M. longissimus thoracis et lumborum transcriptome of steers divergent for residual feed intake.Sci Rep. 2023 Jun 3;13(1):9034. doi: 10.1038/s41598-023-35661-z. Sci Rep. 2023. PMID: 37270611 Free PMC article.
-
Investigation into the effect of divergent feed efficiency phenotype on the bovine rumen microbiota across diet and breed.Sci Rep. 2020 Sep 18;10(1):15317. doi: 10.1038/s41598-020-71458-0. Sci Rep. 2020. PMID: 32948787 Free PMC article.
-
Review: Biological determinants of between-animal variation in feed efficiency of growing beef cattle.Animal. 2018 Dec;12(s2):s321-s335. doi: 10.1017/S1751731118001489. Epub 2018 Aug 24. Animal. 2018. PMID: 30139392 Review.
-
Invited review: Improving feed efficiency of beef cattle - the current state of the art and future challenges.Animal. 2018 Sep;12(9):1815-1826. doi: 10.1017/S1751731118000976. Epub 2018 May 21. Animal. 2018. PMID: 29779496 Review.
Cited by
-
Identification and characterization of circular RNAs in association with the feed efficiency in Hu lambs.BMC Genomics. 2022 Apr 10;23(1):288. doi: 10.1186/s12864-022-08517-5. BMC Genomics. 2022. PMID: 35399048 Free PMC article.
-
Bovine hepatic miRNAome profiling and differential miRNA expression analyses between beef steers with divergent feed efficiency phenotypes.Sci Rep. 2020 Nov 9;10(1):19309. doi: 10.1038/s41598-020-73885-5. Sci Rep. 2020. PMID: 33168877 Free PMC article.
-
Genes associated with body weight gain and feed intake identified by meta-analysis of the mesenteric fat from crossbred beef steers.PLoS One. 2020 Jan 7;15(1):e0227154. doi: 10.1371/journal.pone.0227154. eCollection 2020. PLoS One. 2020. PMID: 31910243 Free PMC article.
-
Carcass Quality Profiles and Associated Genomic Regions of South African Goat Populations Investigated Using Goat SNP50K Genotypes.Animals (Basel). 2022 Feb 2;12(3):364. doi: 10.3390/ani12030364. Animals (Basel). 2022. PMID: 35158687 Free PMC article.
-
An across breed, diet and tissue analysis reveals the transcription factor NR1H3 as a key mediator of residual feed intake in beef cattle.BMC Genomics. 2024 Mar 4;25(1):234. doi: 10.1186/s12864-024-10151-2. BMC Genomics. 2024. PMID: 38438858 Free PMC article.
References
-
- Finneran E, Crosson P, O'Kiely P, Shalloo L, Forristal D, Wallace M. Simulation modelling of the cost of producing and Utilising feeds for ruminants on Irish farms. J Farm Manag. 2010;14(2):95–116.
-
- Berry DP, Crowley JJ. Cell biology symposium: genetics of feed efficiency in dairy and beef cattle. J Anim Sci. 2013;91(4):1594–1613. - PubMed
-
- Gill M, Gibson JP, Lee MRF. Livestock production evolving to contribute to sustainable societies. animal. 2018;12(8):1696–1698. - PubMed
-
- Fitzsimons C, Kenny D, Deighton M, Fahey A, McGee M. Methane emissions, body composition, and rumen fermentation traits of beef heifers differing in residual feed intake. J Anim Sci. 2013;91(12):5789–5800. - PubMed
-
- Archer JA, Richardson EC, Herd RM, Arthur PF. Potential for selection to improve efficiency of feed use in beef cattle: a review. Aust J Agric Res. 1999;50(2):147–162.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

