Using imputed whole-genome sequence data to improve the accuracy of genomic prediction for parasite resistance in Australian sheep
- PMID: 31242855
- PMCID: PMC6595562
- DOI: 10.1186/s12711-019-0476-4
Using imputed whole-genome sequence data to improve the accuracy of genomic prediction for parasite resistance in Australian sheep
Abstract
Background: This study aimed at (1) comparing the accuracies of genomic prediction for parasite resistance in sheep based on whole-genome sequence (WGS) data to those based on 50k and high-density (HD) single nucleotide polymorphism (SNP) panels; (2) investigating whether the use of variants within quantitative trait loci (QTL) regions that were selected from regional heritability mapping (RHM) in an independent dataset improved the accuracy more than variants selected from genome-wide association studies (GWAS); and (3) comparing the prediction accuracies between variants selected from WGS data to variants selected from the HD SNP panel.
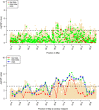
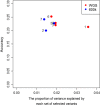
Results: The accuracy of genomic prediction improved marginally from 0.16 ± 0.02 and 0.18 ± 0.01 when using all the variants from 50k and HD genotypes, respectively, to 0.19 ± 0.01 when using all the variants from WGS data. Fitting a GRM from the selected variants alongside a GRM from the 50k SNP genotypes improved the prediction accuracy substantially compared to fitting the 50k SNP genotypes alone. The gain in prediction accuracy was slightly more pronounced when variants were selected from WGS data compared to when variants were selected from the HD panel. When sequence variants that passed the GWAS [Formula: see text] threshold of 3 across the entire genome were selected, the prediction accuracy improved by 5% (up to 0.21 ± 0.01), whereas when selection was limited to sequence variants that passed the same GWAS [Formula: see text] threshold of 3 in regions identified by RHM, the accuracy improved by 9% (up to 0.25 ± 0.01).
Conclusions: Our results show that through careful selection of sequence variants from the QTL regions, the accuracy of genomic prediction for parasite resistance in sheep can be improved. These findings have important implications for genomic prediction in sheep.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Lane J, Jubb T, Shepherd R, Webb-Ware J, Fordyce G. Priority list of endemic diseases for the red meat industries. Final Report. St Lucia: Meat & Livestock Australia L; 2015.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

