Diversity and distribution of type A influenza viruses: an updated panorama analysis based on protein sequences
- PMID: 31242907
- PMCID: PMC6595669
- DOI: 10.1186/s12985-019-1188-7
Diversity and distribution of type A influenza viruses: an updated panorama analysis based on protein sequences
Abstract
Background: Type A influenza viruses (IAVs) cause significant infections in humans and multiple species of animals including pigs, horses, birds, dogs and some marine animals. They are of complicated phylogenetic diversity and distribution, and analysis of their phylogenetic diversity and distribution from a panorama view has not been updated for multiple years.
Methods: 139,872 protein sequences of IAVs from GenBank were selected, and they were aligned and phylogenetically analyzed using the software tool MEGA 7.0. Lineages and subordinate lineages were classified according to the topology of the phylogenetic trees and the host, temporal and spatial distribution of the viruses, and designated using a novel universal nomenclature system.
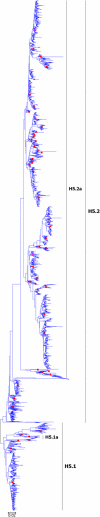
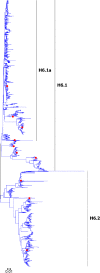
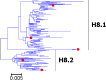
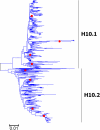
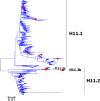
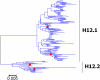
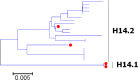
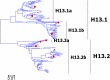
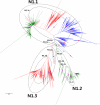
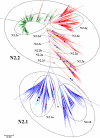
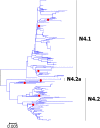
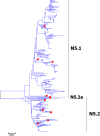
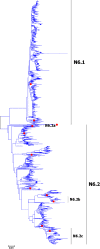
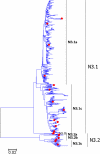
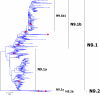
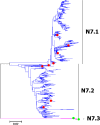
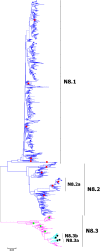
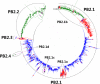
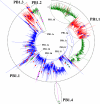
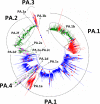
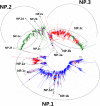
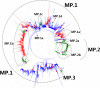
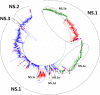
Results: Large phylogenetic trees of the two external viral genes (HA and NA) and six internal genes (PB2, PB1, PA, NP, MP and NS) were constructed, and the diversity and the host, temporal and spatial distribution of these genes were calculated and statistically analyzed. Various features regarding the diversity and distribution of IAVs were confirmed, revised or added through this study, as compared with previous reports. Lineages and subordinate lineages were classified and designated for each of the genes based on the updated panorama views.
Conclusions: The panorama views of phylogenetic diversity and distribution of IAVs and their nomenclature system were updated and assumed to be of significance for studies and communication of IAVs.
Keywords: Distribution; Diversity; Influenza virus; Nomenclature; Phylogenetics; Protein.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Panorama phylogenetic diversity and distribution of type A influenza viruses based on their six internal gene sequences.Virol J. 2009 Sep 8;6:137. doi: 10.1186/1743-422X-6-137. Virol J. 2009. PMID: 19737421 Free PMC article.
-
Panorama phylogenetic diversity and distribution of Type A influenza virus.PLoS One. 2009;4(3):e5022. doi: 10.1371/journal.pone.0005022. Epub 2009 Mar 27. PLoS One. 2009. PMID: 19325912 Free PMC article.
-
The panorama of the diversity of H5 subtype influenza viruses.Virus Genes. 2007 Jun;34(3):283-7. doi: 10.1007/s11262-006-0018-3. Epub 2006 Aug 19. Virus Genes. 2007. PMID: 16924425
-
A Phylogeny-Based Global Nomenclature System and Automated Annotation Tool for H1 Hemagglutinin Genes from Swine Influenza A Viruses.mSphere. 2016 Dec 14;1(6):e00275-16. doi: 10.1128/mSphere.00275-16. eCollection 2016 Nov-Dec. mSphere. 2016. PMID: 27981236 Free PMC article.
-
Isolation and genetic characterization of avian-like H1N1 and novel ressortant H1N2 influenza viruses from pigs in China.Biochem Biophys Res Commun. 2009 Aug 21;386(2):278-83. doi: 10.1016/j.bbrc.2009.05.056. Epub 2009 May 19. Biochem Biophys Res Commun. 2009. PMID: 19460353 Review.
Cited by
-
Design, Synthesis, Biological Evaluation and In Silico Studies of Pyrazole-Based NH2-Acyl Oseltamivir Analogues as Potent Neuraminidase Inhibitors.Pharmaceuticals (Basel). 2021 Apr 16;14(4):371. doi: 10.3390/ph14040371. Pharmaceuticals (Basel). 2021. PMID: 33923858 Free PMC article.
-
Influenza A Virus Hemagglutinin Trimer, Head and Stem Proteins Identify and Quantify Different Hemagglutinin-Specific B Cell Subsets in Humans.Vaccines (Basel). 2021 Jul 2;9(7):717. doi: 10.3390/vaccines9070717. Vaccines (Basel). 2021. PMID: 34358138 Free PMC article.
-
That H9N2 avian influenza viruses circulating in different regions gather in the same live-poultry market poses a potential threat to public health.Front Microbiol. 2023 Feb 16;14:1128286. doi: 10.3389/fmicb.2023.1128286. eCollection 2023. Front Microbiol. 2023. PMID: 36876085 Free PMC article.
-
Multi-COBRA hemagglutinin formulated with cGAMP microparticles elicits protective immune responses against influenza viruses.mSphere. 2024 Jul 30;9(7):e0016024. doi: 10.1128/msphere.00160-24. Epub 2024 Jun 26. mSphere. 2024. PMID: 38920382 Free PMC article.
-
Improved Subtyping of Avian Influenza Viruses Using an RT-qPCR-Based Low Density Array: 'Riems Influenza a Typing Array', Version 2 (RITA-2).Viruses. 2022 Feb 17;14(2):415. doi: 10.3390/v14020415. Viruses. 2022. PMID: 35216008 Free PMC article.
References
-
- Snyder MH, Buckler-White AJ, London WT, Tierney EL, Murphy BR. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian-human influenza A/pintail/79 reassortant viruses for monkeys. J Virol. 1987;61(9):2857–63. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

