Comparative Proteome Profiling and Mutant Protein Identification in Metastatic Prostate Cancer Cells by Quantitative Mass Spectrometry-based Proteogenomics
- PMID: 31243108
- PMCID: PMC6609260
- DOI: 10.21873/cgp.20132
Comparative Proteome Profiling and Mutant Protein Identification in Metastatic Prostate Cancer Cells by Quantitative Mass Spectrometry-based Proteogenomics
Abstract
Background/aim: Prostate cancer (PCa) is the most frequent cancer found in males worldwide. The aim of this study was to identify new biomarkers using mutated peptides for the prognosis and prediction of advanced PCa, based on proteogenomics.
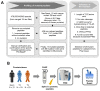
Materials and methods: The tryptic peptides were analyzed by tandem mass tag-based quantitative proteomics. Proteogenomics were used to identify mutant peptides as novel biomarkers in advanced PCa.
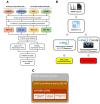
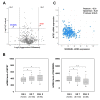
Results: Using a human database, increased levels of INTS7 and decreased levels of SH3BGRL were found to be associated with the aggressiveness of PCa. Using proteogenomics and a cancer mutation database, 70 mutant peptides were identified in PCa cell lines. Using parallel reaction monitoring, the expression of seven mutant peptides was found to be altered in tumors, amongst which CAPN2 D22E was the most significantly up-regulated mutant peptide in PCa tissues.
Conclusion: Altered mutant peptides present in PCa tissue could be used as new biomarkers in advanced PCa.
Keywords: Prostate cancer; metastasis; mutated peptides; proteogenomics; quantitative proteomics.
Copyright© 2019, International Institute of Anticancer Research (Dr. George J. Delinasios), All rights reserved.
Conflict of interest statement
The Authors report no conflicts of interest. The Authors alone are responsible for the content and writing of this article.
Figures



Similar articles
-
Comparative Secretome Profiling and Mutant Protein Identification in Metastatic Prostate Cancer Cells by Quantitative Mass Spectrometry-based Proteomics.Cancer Genomics Proteomics. 2018 Jul-Aug;15(4):279-290. doi: 10.21873/cgp.20086. Cancer Genomics Proteomics. 2018. PMID: 29976633 Free PMC article.
-
Mutant Proteogenomics.Adv Exp Med Biol. 2016;926:77-91. doi: 10.1007/978-3-319-42316-6_6. Adv Exp Med Biol. 2016. PMID: 27686807 Review.
-
The Proteome of Primary Prostate Cancer.Eur Urol. 2016 May;69(5):942-52. doi: 10.1016/j.eururo.2015.10.053. Epub 2015 Dec 2. Eur Urol. 2016. PMID: 26651926
-
Variant peptide detection utilizing mass spectrometry: laying the foundations for proteogenomic identification and validation.Clin Chem Lab Med. 2017 Aug 28;55(9):1291-1304. doi: 10.1515/cclm-2016-0947. Clin Chem Lab Med. 2017. PMID: 28157690
-
Current Challenges and Implications of Proteogenomic Approaches in Prostate Cancer.Curr Top Med Chem. 2020;20(22):1968-1980. doi: 10.2174/1568026620666200722112450. Curr Top Med Chem. 2020. PMID: 32703135 Review.
Cited by
-
Managing a Large-Scale Multiomics Project: A Team Science Case Study in Proteogenomics.Methods Mol Biol. 2021;2194:187-221. doi: 10.1007/978-1-0716-0849-4_11. Methods Mol Biol. 2021. PMID: 32926368 Free PMC article.
-
Characterization of Novel Progression Factors in Castration-Resistant Prostate Cancer Based on Global Comparative Proteome Analysis.Cancers (Basel). 2021 Jul 8;13(14):3432. doi: 10.3390/cancers13143432. Cancers (Basel). 2021. PMID: 34298646 Free PMC article.
-
Prognostic implication of heterogeneity and trajectory progression induced by enzalutamide in prostate cancer.Front Endocrinol (Lausanne). 2023 Mar 16;14:1148898. doi: 10.3389/fendo.2023.1148898. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37008945 Free PMC article.
-
Mutant p53 induces SH3BGRL expression to promote cell engulfment.Cell Death Discov. 2025 Jul 1;11(1):288. doi: 10.1038/s41420-025-02582-x. Cell Death Discov. 2025. PMID: 40592808 Free PMC article.
-
Prostate cancer in omics era.Cancer Cell Int. 2022 Sep 5;22(1):274. doi: 10.1186/s12935-022-02691-y. Cancer Cell Int. 2022. PMID: 36064406 Free PMC article.
References
-
- Wang Q, Chaerkady R, Wu J, Hwang HJ, Papadopoulos N, Kopelovich L, Maitra A, Matthaei H, Eshleman JR, Hruban RH, Kinzler KW, Pandey A, Vogelstein B. Mutant proteins as cancer-specific biomarkers. Proc Natl Acad Sci USA. 2011;108(6):2444–2449. PMID: 21248225. DOI: 10.1073/pnas.1019 203108. - PMC - PubMed
-
- American Cancer Society: Cancer facts & figures. The Society. 2018. Available from: https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
