The role of gelsolin domain 3 in familial amyloidosis (Finnish type)
- PMID: 31243148
- PMCID: PMC6628662
- DOI: 10.1073/pnas.1902189116
The role of gelsolin domain 3 in familial amyloidosis (Finnish type)
Abstract
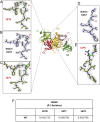
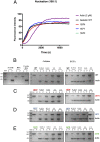
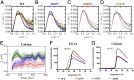
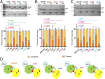
In the disease familial amyloidosis, Finnish type (FAF), also known as AGel amyloidosis (AGel), the mechanism by which point mutations in the calcium-regulated actin-severing protein gelsolin lead to furin cleavage is not understood in the intact protein. Here, we provide a structural and biochemical characterization of the FAF variants. X-ray crystallography structures of the FAF mutant gelsolins demonstrate that the mutations do not significantly disrupt the calcium-free conformations of gelsolin. Small-angle X-ray-scattering (SAXS) studies indicate that the FAF calcium-binding site mutants are slower to activate, whereas G167R is as efficient as the wild type. Actin-regulating studies of the gelsolins at the furin cleavage pH (6.5) show that the mutant gelsolins are functional, suggesting that they also adopt relatively normal active conformations. Deletion of gelsolin domains leads to sensitization to furin cleavage, and nanobody-binding protects against furin cleavage. These data indicate instability in the second domain of gelsolin (G2), since loss or gain of G2-stabilizing interactions impacts the efficiency of cleavage by furin. To demonstrate this principle, we engineered non-FAF mutations in G3 that disrupt the G2-G3 interface in the calcium-activated structure. These mutants led to increased furin cleavage. We carried out molecular dynamics (MD) simulations on the FAF and non-FAF mutant G2-G3 fragments of gelsolin. All mutants showed an increase in the distance between the center of masses of the 2 domains (G2 and G3). Since G3 covers the furin cleavage site on G2 in calcium-activated gelsolin, this suggests that destabilization of this interface is a critical step in cleavage.
Keywords: AGel amyloidosis; FAF; amyloid; gelsolin; structure.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.Biochim Biophys Acta Mol Basis Dis. 2019 Mar 1;1865(3):648-660. doi: 10.1016/j.bbadis.2019.01.010. Epub 2019 Jan 6. Biochim Biophys Acta Mol Basis Dis. 2019. PMID: 30625383
-
Gelsolin domain 2 Ca2+ affinity determines susceptibility to furin proteolysis and familial amyloidosis of finnish type.J Mol Biol. 2003 Nov 14;334(1):119-27. doi: 10.1016/j.jmb.2003.09.029. J Mol Biol. 2003. PMID: 14596804
-
Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF.EMBO J. 2004 Jul 21;23(14):2713-22. doi: 10.1038/sj.emboj.7600280. Epub 2004 Jun 24. EMBO J. 2004. PMID: 15215896 Free PMC article.
-
Ca2+ binding protects against gelsolin amyloidosis.Biochem Biophys Res Commun. 2004 Oct 1;322(4):1105-10. doi: 10.1016/j.bbrc.2004.07.125. Biochem Biophys Res Commun. 2004. PMID: 15336957 Review.
-
Gelsolin amyloidosis: genetics, biochemistry, pathology and possible strategies for therapeutic intervention.Crit Rev Biochem Mol Biol. 2012 May-Jun;47(3):282-96. doi: 10.3109/10409238.2012.661401. Epub 2012 Feb 24. Crit Rev Biochem Mol Biol. 2012. PMID: 22360545 Free PMC article. Review.
Cited by
-
Inverse Boltzmann Iterative Multi-Scale Molecular Dynamics Study between Carbon Nanotubes and Amino Acids.Molecules. 2022 Apr 27;27(9):2785. doi: 10.3390/molecules27092785. Molecules. 2022. PMID: 35566140 Free PMC article.
-
Molecular Dynamics Study of the Curvature-Driven Interactions between Carbon-Based Nanoparticles and Amino Acids.Molecules. 2023 Jan 4;28(2):482. doi: 10.3390/molecules28020482. Molecules. 2023. PMID: 36677540 Free PMC article.
-
The structure of N184K amyloidogenic variant of gelsolin highlights the role of the H-bond network for protein stability and aggregation properties.Eur Biophys J. 2020 Jan;49(1):11-19. doi: 10.1007/s00249-019-01409-9. Epub 2019 Nov 13. Eur Biophys J. 2020. PMID: 31724080
-
Analyses Mutations in GSN, CST3, TTR, and ITM2B Genes in Chinese Patients With Alzheimer's Disease.Front Aging Neurosci. 2020 Sep 10;12:581524. doi: 10.3389/fnagi.2020.581524. eCollection 2020. Front Aging Neurosci. 2020. PMID: 33192475 Free PMC article.
-
Edge Strand Dissociation and Conformational Changes in Transthyretin under Amyloidogenic Conditions.Biophys J. 2020 Nov 17;119(10):1995-2009. doi: 10.1016/j.bpj.2020.08.043. Epub 2020 Oct 20. Biophys J. 2020. PMID: 33091379 Free PMC article.
References
-
- Kiuru S., Matikainen E., Kupari M., Haltia M., Palo J., Autonomic nervous system and cardiac involvement in familial amyloidosis, Finnish type (FAF). J. Neurol. Sci. 126, 40–48 (1994). - PubMed
-
- Maury C. P., Kere J., Tolvanen R., de la Chapelle A., Homozygosity for the Asn187 gelsolin mutation in Finnish-type familial amyloidosis is associated with severe renal disease. Genomics 13, 902–903 (1992). - PubMed
-
- Meretoja J., Familial systemic paramyloidosis with lattice dystrophy of the cornea, progressive cranial neuropathy, skin changes and various internal symptoms. A previously unrecognized heritable syndrome. Ann. Clin. Res. 1, 314–324 (1969). - PubMed
-
- Taira M., et al. , Clinical features and haplotype analysis of newly identified Japanese patients with gelsolin-related familial amyloidosis of Finnish type. Neurogenetics 13, 237–243 (2012). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Research Materials

