DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation
- PMID: 31243293
- PMCID: PMC6594956
- DOI: 10.1038/s41467-019-10844-3
DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation
Abstract
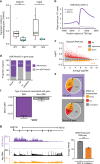
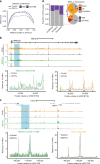
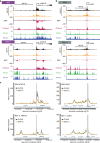
Enhancer elements are a key regulatory feature of many important genes. Several general features including the presence of specific histone modifications are used to demarcate potentially active enhancers. Here we reveal that putative enhancers marked with H3 lysine 79 (H3K79) di or trimethylation (me2/3) (which we name H3K79me2/3 enhancer elements or KEEs) can be found in multiple cell types. Mixed lineage leukemia gene (MLL) rearrangements (MLL-r) such as MLL-AF4 are a major cause of incurable acute lymphoblastic leukemias (ALL). Using the DOT1L inhibitor EPZ-5676 in MLL-AF4 leukemia cells, we show that H3K79me2/3 is required for maintaining chromatin accessibility, histone acetylation and transcription factor binding specifically at KEEs but not non-KEE enhancers. We go on to show that H3K79me2/3 is essential for maintaining enhancer-promoter interactions at a subset of KEEs. Together, these data implicate H3K79me2/3 as having a functional role at a subset of active enhancers in MLL-AF4 leukemia cells.
Conflict of interest statement
P.V. and T.A.M. are founding shareholders of Oxstem Oncology (OSO), a subsidiary company of OxStem Ltd. The other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_12009/15/MRC_/Medical Research Council/United Kingdom
- MR/L008963/1/MRC_/Medical Research Council/United Kingdom
- MC_PC_15065/MRC_/Medical Research Council/United Kingdom
- MC_UU_12009/11/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- G1000729/MRC_/Medical Research Council/United Kingdom
- MC_UU_12009/6/MRC_/Medical Research Council/United Kingdom
- MC_UU_00016/6/MRC_/Medical Research Council/United Kingdom
- MC_UU_00016/11/MRC_/Medical Research Council/United Kingdom
- MC_U137961146/MRC_/Medical Research Council/United Kingdom
- MC_UU_00016/14/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

