Developmental dynamics of lncRNAs across mammalian organs and species
- PMID: 31243368
- PMCID: PMC6660317
- DOI: 10.1038/s41586-019-1341-x
Developmental dynamics of lncRNAs across mammalian organs and species
Abstract
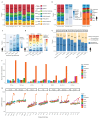
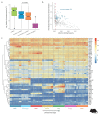
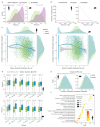
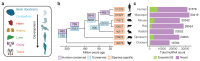
Although many long noncoding RNAs (lncRNAs) have been identified in human and other mammalian genomes, there has been limited systematic functional characterization of these elements. In particular, the contribution of lncRNAs to organ development remains largely unexplored. Here we analyse the expression patterns of lncRNAs across developmental time points in seven major organs, from early organogenesis to adulthood, in seven species (human, rhesus macaque, mouse, rat, rabbit, opossum and chicken). Our analyses identified approximately 15,000 to 35,000 candidate lncRNAs in each species, most of which show species specificity. We characterized the expression patterns of lncRNAs across developmental stages, and found many with dynamic expression patterns across time that show signatures of enrichment for functionality. During development, there is a transition from broadly expressed and conserved lncRNAs towards an increasing number of lineage- and organ-specific lncRNAs. Our study provides a resource of candidate lncRNAs and their patterns of expression and evolutionary conservation across mammalian organ development.
Conflict of interest statement
The authors declare no competing financial interests.
Figures









References
-
- Carninci P, et al. The Transcriptional Landscape of the Mammalian Genome. Science (80-. ) 2005;309:1559–1563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

