FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes
- PMID: 31243370
- PMCID: PMC6661172
- DOI: 10.1038/s41586-019-1318-9
FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes
Erratum in
-
Author Correction: FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes.Nature. 2020 Sep;585(7826):E20. doi: 10.1038/s41586-020-2678-x. Nature. 2020. PMID: 32895534
Abstract
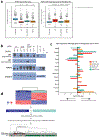
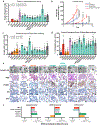
Mutations in the transcription factor FOXA1 define a unique subset of prostate cancers but the functional consequences of these mutations and whether they confer gain or loss of function is unknown1-9. Here, by annotating the landscape of FOXA1 mutations from 3,086 human prostate cancers, we define two hotspots in the forkhead domain: Wing2 (around 50% of all mutations) and the highly conserved DNA-contact residue R219 (around 5% of all mutations). Wing2 mutations are detected in adenocarcinomas at all stages, whereas R219 mutations are enriched in metastatic tumours with neuroendocrine histology. Interrogation of the biological properties of wild-type FOXA1 and fourteen FOXA1 mutants reveals gain of function in mouse prostate organoid proliferation assays. Twelve of these mutants, as well as wild-type FOXA1, promoted an exaggerated pro-luminal differentiation program, whereas two different R219 mutants blocked luminal differentiation and activated a mesenchymal and neuroendocrine transcriptional program. Assay for transposase-accessible chromatin using sequencing (ATAC-seq) of wild-type FOXA1 and representative Wing2 and R219 mutants revealed marked, mutant-specific changes in open chromatin at thousands of genomic loci and exposed sites of FOXA1 binding and associated increases in gene expression. Of note, ATAC-seq peaks in cells expressing R219 mutants lacked the canonical core FOXA1-binding motifs (GTAAAC/T) but were enriched for a related, non-canonical motif (GTAAAG/A), which was preferentially activated by R219-mutant FOXA1 in reporter assays. Thus, FOXA1 mutations alter its pioneering function and perturb normal luminal epithelial differentiation programs, providing further support for the role of lineage plasticity in cancer progression.
Figures












Comment in
-
The Sly Oncogene: FOXA1 Mutations in Prostate Cancer.Cancer Cell. 2019 Aug 12;36(2):119-121. doi: 10.1016/j.ccell.2019.07.005. Cancer Cell. 2019. PMID: 31408618
References
Main Text References
Methods References
-
- Cerami E et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery 2, 401, doi: 10.1158/2159-8290.CD-12-0095 (2012). - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 CA160001/CA/NCI NIH HHS/United States
- R01 CA193837/CA/NCI NIH HHS/United States
- R01 CA233650/CA/NCI NIH HHS/United States
- P50 CA092629/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- K08 CA226348/CA/NCI NIH HHS/United States
- U54 CA224079/CA/NCI NIH HHS/United States
- T32 GM083937/GM/NIGMS NIH HHS/United States
- R37 CA215040/CA/NCI NIH HHS/United States
- F99 CA223063/CA/NCI NIH HHS/United States
- R01 CA155169/CA/NCI NIH HHS/United States
- K08 CA187417/CA/NCI NIH HHS/United States
- P50 CA211024/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

