NMR and MD studies combined to elucidate inhibitor and water interactions of HIV-1 protease and their modulations with resistance mutations
- PMID: 31243634
- PMCID: PMC6941145
- DOI: 10.1007/s10858-019-00260-6
NMR and MD studies combined to elucidate inhibitor and water interactions of HIV-1 protease and their modulations with resistance mutations
Abstract
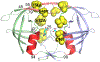
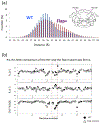
Over the last two decades, both the sensitivity of NMR and the time scale of molecular dynamics (MD) simulation have increased tremendously and have advanced the field of protein dynamics. HIV-1 protease has been extensively studied using these two methods, and has presented a framework for cross-evaluation of structural ensembles and internal dynamics by integrating the two methods. Here, we review studies from our laboratories over the last several years, to understand the mechanistic basis of protease drug-resistance mutations and inhibitor responses, using NMR and crystal structure-based parallel MD simulations. Our studies demonstrate that NMR relaxation experiments, together with crystal structures and MD simulations, significantly contributed to the current understanding of structural/dynamic changes due to HIV-1 protease drug resistance mutations.
Keywords: Crystal structures; Drug design; HIV-1; Inhibitor; MD; NMR; Protease.
Figures


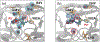
References
-
- Altman MD, Ali A, Reddy GS, Nalam MN, Anjum SG, Cao H, Chellappan S, Kairys V, Fernandes MX, Gilson MK, Schiffer CA, Rana TM & Tidor B (2008). HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants. J Am Chem Soc 130: 6099–113. - PMC - PubMed
-
- Baldwin ET, Bhat TN, Gulnik S, Liu B, Topol IA, Kiso Y, Mimoto T, Mitsuya H & Erickson JW (1995). Structure of HIV-1 protease with KNI-272, a tight-binding transition-state analog containing allophenylnorstatine. Structure 3: 581–590. - PubMed
-
- Bandyopadhyay P & Meher BR (2006). Drug resistance of HIV-1 protease against JE-2147: I47V mutation investigated by molecular dynamics simulation. Chem Biol Drug Des 67: 155–61. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

