Light-induced complex formation of bacteriophytochrome RpBphP1 and gene repressor RpPpsR2 probed by SAXS
- PMID: 31243889
- PMCID: PMC6899989
- DOI: 10.1111/febs.14973
Light-induced complex formation of bacteriophytochrome RpBphP1 and gene repressor RpPpsR2 probed by SAXS
Abstract
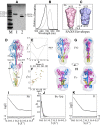
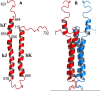
Bacteriophytochrome proteins (BphPs) are molecular light switches that enable organisms to adapt to changing light conditions through the control of gene expression. Canonical type 1 BphPs have histidine kinase output domains, but type 3 RpBphP1, in the bacterium Rhodopseudomonas palustris (Rps. palustris), has a C terminal PAS9 domain and a two-helix output sensor (HOS) domain. Type 1 BphPs form head-to-head parallel dimers; however, the crystal structure of RpBphP1ΔHOS, which does not contain the HOS domain, revealed pseudo anti-parallel dimers. HOS domains are homologs of Dhp dimerization domains in type 1 BphPs. We show, by applying the small angle X-ray scattering (SAXS) technique on full-length RpBphP1, that HOS domains fulfill a similar role in the formation of parallel dimers. On illumination with far-red light, RpBphP1 forms a complex with gene repressor RpPpsR2 through light-induced structural changes in its HOS domains. An RpBphP1:RpPpsR2 complex is formed in the molecular ratio of 2 : 1 such that one RpBphP1 dimer binds one RpPpsR2 monomer. Molecular dimers have been modeled with Pfr and Pr SAXS data, suggesting that, in the Pfr state, stable dimeric four α-helix bundles are formed between HOS domains, rendering RpBphP1functionally inert. On illumination with light of 760 nm wavelength, four α-helix bundles formed by HOS dimers are disrupted, rendering helices available for binding with RpPpsR2.
Keywords: SAXS; bacteriophytochrome; complex formation; photo-induced changes; photosynthesis.
© 2019 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor.Structure. 2012 Aug 8;20(8):1436-46. doi: 10.1016/j.str.2012.06.002. Epub 2012 Jul 12. Structure. 2012. PMID: 22795083
-
Small-angle X-ray scattering reveals the solution structure of a bacteriophytochrome in the catalytically active Pr state.J Mol Biol. 2006 Dec 8;364(4):655-66. doi: 10.1016/j.jmb.2006.09.045. Epub 2006 Sep 23. J Mol Biol. 2006. PMID: 17027028
-
Structure of the Full-Length Bacteriophytochrome from the Plant Pathogen Xanthomonas campestris Provides Clues to its Long-Range Signaling Mechanism.J Mol Biol. 2016 Sep 25;428(19):3702-20. doi: 10.1016/j.jmb.2016.04.012. Epub 2016 Apr 20. J Mol Biol. 2016. PMID: 27107635
-
The Crystal Structures of the N-terminal Photosensory Core Module of Agrobacterium Phytochrome Agp1 as Parallel and Anti-parallel Dimers.J Biol Chem. 2016 Sep 23;291(39):20674-91. doi: 10.1074/jbc.M116.739136. Epub 2016 Jul 26. J Biol Chem. 2016. PMID: 27466363 Free PMC article.
-
Photoactive yellow protein, bacteriophytochrome, and sensory rhodopsin in purple phototrophic bacteria.Photochem Photobiol Sci. 2004 Jun;3(6):519-30. doi: 10.1039/b315731h. Epub 2004 Apr 20. Photochem Photobiol Sci. 2004. PMID: 15170480 Review.
Cited by
-
Engineering Photosensory Modules of Non-Opsin-Based Optogenetic Actuators.Int J Mol Sci. 2020 Sep 7;21(18):6522. doi: 10.3390/ijms21186522. Int J Mol Sci. 2020. PMID: 32906617 Free PMC article. Review.
-
Red light-induced structure changes in phytochrome A from Pisum sativum.Sci Rep. 2021 Feb 2;11(1):2827. doi: 10.1038/s41598-021-82544-2. Sci Rep. 2021. PMID: 33531580 Free PMC article.
-
Traits of Bathy Phytochromes and Application to Bacterial Optogenetics.ACS Synth Biol. 2025 Aug 15;14(8):3207-3218. doi: 10.1021/acssynbio.5c00337. Epub 2025 Jul 11. ACS Synth Biol. 2025. PMID: 40645610 Free PMC article.
References
-
- Giraud E, Fardoux J, Fourrier N, Hannibal L, Genty B, Bouyer P, Dreyfus B & Vermeglio A (2002) Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417, 202–205. - PubMed
-
- Bellini D & Papiz MZ (2012) Structure of a bacteriophytochrome and light‐stimulated protomer swapping with a gene repressor. Structure 20, 1436–1446. - PubMed
-
- Braatsch S, Johnson JA, Noll K & Beatty JT (2007) The O2‐responsive repressor PpsR2 but not PpsR1 transduces a light signal sensed by the BphP1 phytochrome in Rhodopseudomonas palustris CGA009. FEMS Microbiol Lett 272, 60–64. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

