MEA6 Deficiency Impairs Cerebellar Development and Motor Performance by Tethering Protein Trafficking
- PMID: 31244610
- PMCID: PMC6580151
- DOI: 10.3389/fncel.2019.00250
MEA6 Deficiency Impairs Cerebellar Development and Motor Performance by Tethering Protein Trafficking
Abstract
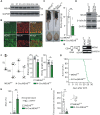
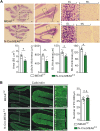
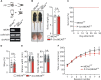
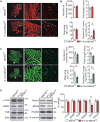
Meningioma expressed antigen 6 (MEA6), also called cutaneous T cell lymphoma-associated antigen 5 (cTAGE5), was initially found in tumor tissues. MEA6 is located in endoplasmic reticulum (ER) exit sites and regulates the transport of collagen, very low density lipoprotein, and insulin. It is also reported that MEA6 might be related to Fahr's syndrome, which comprises neurological, movement, and neuropsychiatric disorders. Here, we show that MEA6 is critical to cerebellar development and motor performance. Mice with conditional knockout of MEA6 (Nestin-Cre;MEA6F/F) display smaller sizes of body and brain compared to control animals, and survive maximal 28 days after birth. Immunohistochemical and behavioral studies demonstrate that these mutant mice have defects in cerebellar development and motor performance. In contrast, PC deletion of MEA6 (pCP2-Cre;MEA6F/F) causes milder phenotypes in cerebellar morphology and motor behaviors. While pCP2-Cre;MEA6F/F mice have normal lobular formation and gait, they present the extensive self-crossing of PC dendrites and damaged motor learning. Interestingly, the expression of key molecules that participates in cerebellar development, including Slit2 and brain derived neurotrophic factor (BDNF), is significantly increased in ER, suggesting that MEA6 ablation impairs ER function and thus these proteins are arrested in ER. Our study provides insight into the roles of MEA6 in the brain and the pathogenesis of Fahr's syndrome.
Keywords: Fahr’s syndrome; MEA6; Slit2; dendrite self-crossing; motor learning.
Figures


Comment in
-
Digenic Variants as Possible Clinical Modifier of Primary Familial Brain Calcification Patients.J Mol Neurosci. 2020 Jan;70(1):142-144. doi: 10.1007/s12031-019-01430-9. Epub 2019 Nov 25. J Mol Neurosci. 2020. PMID: 31768941
Similar articles
-
Deletion of Mea6 in Cerebellar Granule Cells Impairs Synaptic Development and Motor Performance.Front Cell Dev Biol. 2021 Feb 25;8:627146. doi: 10.3389/fcell.2020.627146. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33718348 Free PMC article.
-
cTAGE5/MEA6 plays a critical role in neuronal cellular components trafficking and brain development.Proc Natl Acad Sci U S A. 2018 Oct 2;115(40):E9449-E9458. doi: 10.1073/pnas.1804083115. Epub 2018 Sep 17. Proc Natl Acad Sci U S A. 2018. PMID: 30224460 Free PMC article.
-
cTAGE5 deletion in pancreatic β cells impairs proinsulin trafficking and insulin biogenesis in mice.J Cell Biol. 2017 Dec 4;216(12):4153-4164. doi: 10.1083/jcb.201705027. Epub 2017 Nov 13. J Cell Biol. 2017. PMID: 29133483 Free PMC article.
-
Fahr's syndrome due to hypoparathyroidism revisited: A case of parkinsonism and a review of all published cases.Clin Neurol Neurosurg. 2021 Mar;202:106514. doi: 10.1016/j.clineuro.2021.106514. Epub 2021 Jan 22. Clin Neurol Neurosurg. 2021. PMID: 33529967 Review.
-
Fahr's syndrome: literature review of current evidence.Orphanet J Rare Dis. 2013 Oct 8;8:156. doi: 10.1186/1750-1172-8-156. Orphanet J Rare Dis. 2013. PMID: 24098952 Free PMC article. Review.
Cited by
-
Sepsis Impairs Purkinje Cell Functions and Motor Behaviors Through Microglia Activation.Cerebellum. 2024 Apr;23(2):329-339. doi: 10.1007/s12311-023-01531-7. Epub 2023 Feb 15. Cerebellum. 2024. PMID: 36790600
-
Digenic Variants as Possible Clinical Modifier of Primary Familial Brain Calcification Patients.J Mol Neurosci. 2020 Jan;70(1):142-144. doi: 10.1007/s12031-019-01430-9. Epub 2019 Nov 25. J Mol Neurosci. 2020. PMID: 31768941
-
Deletion of Mea6 in Cerebellar Granule Cells Impairs Synaptic Development and Motor Performance.Front Cell Dev Biol. 2021 Feb 25;8:627146. doi: 10.3389/fcell.2020.627146. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33718348 Free PMC article.
-
KIF2C regulates synaptic plasticity and cognition in mice through dynamic microtubule depolymerization.Elife. 2022 Feb 9;11:e72483. doi: 10.7554/eLife.72483. Elife. 2022. PMID: 35138249 Free PMC article.
-
Ablation of Mea6/cTAGE5 in oligodendrocytes significantly impairs white matter structure and lipid content.Life Metab. 2023 Mar 23;2(2):load010. doi: 10.1093/lifemeta/load010. eCollection 2023 Apr. Life Metab. 2023. PMID: 39872732 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

