The Effect of Genetic Variation on the Placental Transcriptome in Humans
- PMID: 31244887
- PMCID: PMC6581026
- DOI: 10.3389/fgene.2019.00550
The Effect of Genetic Variation on the Placental Transcriptome in Humans
Abstract
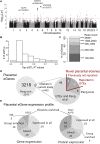
The knowledge of genetic variants shaping human placental transcriptome is limited and they are not cataloged in the Genotype-Tissue Expression project. So far, only one whole genome analysis of placental expression quantitative trait loci (eQTLs) has been published by Peng et al. (2017) with no external independent validation. We report the second study on the landscape of placental eQTLs. The study aimed to generate a high-confidence list of placental cis-eQTLs and to investigate their potential functional implications. Analysis of cis-eQTLs (±100 kbp from the gene) utilized 40 placental RNA sequencing and respective whole genome genotyping datasets. The identified 199 placental cis-eSNPs represented 88 independent eQTL signals (FDR < 5%). The most significant placental eQTLs (FDR < 10-5) modulated the expression of ribosomal protein RPL9, transcription factor ZSCAN9 and aminopeptidase ERAP2. The analysis confirmed 50 eSNP-eGenes pairs reported by Peng et al. (2017) and thus, can be claimed as robust placental eQTL signals. The study identified also 13 novel placental eGenes. Among these, ZSCAN9 is modulated by several eSNPs (experimentally validated: rs1150707) that have been also shown to affect the methylation level of genes variably escaping X-chromosomal inactivation. The identified 63 placental eGenes exhibited mostly mixed or ubiquitous expression. Functional enrichment analysis highlighted 35 Gene Ontology categories with the top ranking pathways "ruffle membrane" (FDR = 1.81 × 10-2) contributing to the formation of motile cell surface and "ATPase activity, coupled" (FDR = 2.88 × 10-2), critical for the membrane transport. Placental eGenes were also significantly enriched in pathways implicated in development, signaling and immune function. However, this study was not able to confirm a significant overrepresentation of genome-wide association studies top hits among the placental eSNP and eGenes, reported by Peng et al. (2017). The identified eSNPs were further analyzed in association with newborn and pregnancy traits. In the discovery step, a suggestive association was detected between the eQTL of ALPG (rs11678251) and reduced placental, newborn's and infant's weight. Meta-analysis across REPROMETA, HAPPY PREGNANCY, ALSPAC cohorts (n = 6830) did not replicate these findings. In summary, the study emphasizes the role of genetic variation in driving the transcriptome profile of the human placenta and the importance to explore further its functional implications.
Keywords: ALSPAC; HAPPY PREGNANCY; REPROMETA; cis-eQTL; complex traits; human placenta.
Figures


Similar articles
-
Current knowledge on genetic variants shaping placental transcriptome and their link to gestational and postnatal health.Placenta. 2021 Dec;116:2-11. doi: 10.1016/j.placenta.2021.02.009. Epub 2021 Feb 22. Placenta. 2021. PMID: 33663810 Review.
-
Genetic Architecture of Gene Expression in European and African Americans: An eQTL Mapping Study in GENOA.Am J Hum Genet. 2020 Apr 2;106(4):496-512. doi: 10.1016/j.ajhg.2020.03.002. Epub 2020 Mar 26. Am J Hum Genet. 2020. PMID: 32220292 Free PMC article.
-
Mapping adipose and muscle tissue expression quantitative trait loci in African Americans to identify genes for type 2 diabetes and obesity.Hum Genet. 2016 Aug;135(8):869-80. doi: 10.1007/s00439-016-1680-8. Epub 2016 May 19. Hum Genet. 2016. PMID: 27193597 Free PMC article.
-
Tissue specific regulation of transcription in endometrium and association with disease.Hum Reprod. 2020 Feb 29;35(2):377-393. doi: 10.1093/humrep/dez279. Hum Reprod. 2020. PMID: 32103259 Free PMC article.
-
Whole genome DNA and RNA sequencing of whole blood elucidates the genetic architecture of gene expression underlying a wide range of diseases.Sci Rep. 2022 Nov 23;12(1):20167. doi: 10.1038/s41598-022-24611-w. Sci Rep. 2022. PMID: 36424512 Free PMC article.
Cited by
-
Genetic variation in placental insufficiency: What have we learned over time?Front Cell Dev Biol. 2022 Oct 14;10:1038358. doi: 10.3389/fcell.2022.1038358. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36313546 Free PMC article. Review.
-
The genetic architecture of sporadic and multiple consecutive miscarriage.Nat Commun. 2020 Nov 25;11(1):5980. doi: 10.1038/s41467-020-19742-5. Nat Commun. 2020. PMID: 33239672 Free PMC article.
-
Coordinated Expressional Landscape of the Human Placental miRNome and Transcriptome.Front Cell Dev Biol. 2021 Jul 21;9:697947. doi: 10.3389/fcell.2021.697947. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34368147 Free PMC article.
-
The Multifaceted Nature of Aminopeptidases ERAP1, ERAP2, and LNPEP: From Evolution to Disease.Front Immunol. 2020 Jul 23;11:1576. doi: 10.3389/fimmu.2020.01576. eCollection 2020. Front Immunol. 2020. PMID: 32793222 Free PMC article.
-
Pan-Genomic Regulation of Gene Expression in Normal and Pathological Human Placentas.Cells. 2023 Feb 10;12(4):578. doi: 10.3390/cells12040578. Cells. 2023. PMID: 36831244 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

