Single-cell RNA sequencing for the study of lupus nephritis
- PMID: 31245017
- PMCID: PMC6560921
- DOI: 10.1136/lupus-2019-000329
Single-cell RNA sequencing for the study of lupus nephritis
Abstract
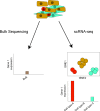
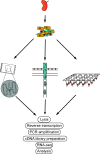
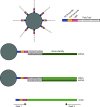
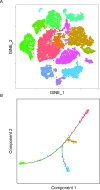
Single-cell RNA sequencing (scRNA-seq) has recently undergone rapid advances in the development of this technology, leading to high throughput and accelerating discovery in many biological systems and diseases. The single-cell resolution of the technique allows for the investigation of heterogeneity in cell populations, and the pinpointing of pathological populations contributing to disease. Here we review the development of scRNA-seq technology and the analysis that has evolved with the ever-increasing throughput. Finally, we highlight recent applications of scRNA-seq to understand the molecular pathogenesis of lupus and lupus nephritis.
Keywords: inflammation; lupus nephritis; systemic lupus erythematosus.
Conflict of interest statement
Competing interests: None declared.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
