Alterations to the Gut Microbiota and Their Correlation With Inflammatory Factors in Chronic Kidney Disease
- PMID: 31245306
- PMCID: PMC6581668
- DOI: 10.3389/fcimb.2019.00206
Alterations to the Gut Microbiota and Their Correlation With Inflammatory Factors in Chronic Kidney Disease
Abstract
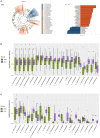
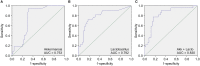
Alterations to the gut microbiota have been previously suggested to be tightly linked to chronic systemic inflammation, which is a major contributing factor to complications and disease progression in chronic kidney disease (CKD). Nevertheless, the effect of gut dysbiosis on the pathogenesis and/or production of inflammatory factors in CKD has not been extensively studied to date. In the present study, we conducted 16S ribosomal DNA pyrosequencing using fecal microbiota samples and analyzed the production of serum inflammatory factors in 50 patients with CKD and 22 healthy control (HC) subjects. The results revealed that compared to the HC subjects, patients with CKD exhibited a significant reduction in the richness and structure of their fecal microbiota. At the phylum level, compared to the HC group, patients with CKD also presented reduced abundance of Actinobacteria but increased abundance of Verrucomicrobia. Moreover, the genera Lactobacillus, Clostridium IV, Paraprevotella, Clostridium sensu stricto, Desulfovibrio, and Alloprevotella were enriched in the fecal samples of patients with CKD, while Akkermansia and Parasutterella were enriched in those of the HC subjects. The abundance of Akkermansia in the CKD group was significantly lower than that in the HC group (3.08 vs. 0.67%); this decrease in the abundance of Akkermansia, an important probiotic, in patients with CKD is a striking discovery as it has not been previously reported. Finally, we analyzed whether these changes to the fecal microbiota correlated with CKD clinical characteristics and/or the production of known inflammatory factors. Altered levels of the microbiota genera Parasutterella, Lactobacillus, Paraprevotella, Clostridium sensu stricto, and Desulfovibrio were shown to be correlated with CKD disease-severity indicators, including the estimated glomerular filtration rate. Most notably, Akkermansia was significantly negatively correlated with the production of interleukin-10. The results of the present study suggest that microbiota dysbiosis may promote chronic systemic inflammation in CKD. Furthermore, they support that modifying the gut microbiota, especially Akkermansia, may be a promising potential therapeutic strategy to attenuate the progression of, and/or systemic inflammation in, CKD.
Keywords: 16S rDNA deep sequencing; Akkermansia; chronic kidney disease; gut microbiota; inflammatory factors.
Figures



Similar articles
-
Characterizing the gut microbiota in patients with chronic kidney disease.Postgrad Med. 2020 Aug;132(6):495-505. doi: 10.1080/00325481.2020.1744335. Epub 2020 Apr 2. Postgrad Med. 2020. PMID: 32241215
-
Disorder of gut amino acids metabolism during CKD progression is related with gut microbiota dysbiosis and metagenome change.J Pharm Biomed Anal. 2018 Feb 5;149:425-435. doi: 10.1016/j.jpba.2017.11.040. Epub 2017 Nov 15. J Pharm Biomed Anal. 2018. PMID: 29169110
-
Uncovering specific taxonomic and functional alteration of gut microbiota in chronic kidney disease through 16S rRNA data.Front Cell Infect Microbiol. 2024 Apr 19;14:1363276. doi: 10.3389/fcimb.2024.1363276. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38707511 Free PMC article.
-
Gut microbiota and chronic kidney disease: evidences and mechanisms that mediate a new communication in the gastrointestinal-renal axis.Pflugers Arch. 2020 Mar;472(3):303-320. doi: 10.1007/s00424-020-02352-x. Epub 2020 Feb 17. Pflugers Arch. 2020. PMID: 32064574 Review.
-
Specific alterations in gut microbiota in patients with chronic kidney disease: an updated systematic review.Ren Fail. 2021 Dec;43(1):102-112. doi: 10.1080/0886022X.2020.1864404. Ren Fail. 2021. PMID: 33406960 Free PMC article.
Cited by
-
Characterization of Fecal Microbiota with Clinical Specimen Using Long-Read and Short-Read Sequencing Platform.Int J Mol Sci. 2020 Sep 26;21(19):7110. doi: 10.3390/ijms21197110. Int J Mol Sci. 2020. PMID: 32993155 Free PMC article.
-
Correlating gut microbial membership to brown bear health metrics.Sci Rep. 2022 Sep 22;12(1):15415. doi: 10.1038/s41598-022-19527-4. Sci Rep. 2022. PMID: 36138067 Free PMC article.
-
Gut Dysbiosis and Kidney Diseases.Front Med (Lausanne). 2022 Mar 3;9:829349. doi: 10.3389/fmed.2022.829349. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35308555 Free PMC article. Review.
-
Gut Mycobiome in Patients With Chronic Kidney Disease Was Altered and Associated With Immunological Profiles.Front Immunol. 2022 Jun 16;13:843695. doi: 10.3389/fimmu.2022.843695. eCollection 2022. Front Immunol. 2022. PMID: 35784313 Free PMC article.
-
Outer membrane vesicles derived from gut microbiota mediate tubulointerstitial inflammation: a potential new mechanism for diabetic kidney disease.Theranostics. 2023 Jul 9;13(12):3988-4003. doi: 10.7150/thno.84650. eCollection 2023. Theranostics. 2023. PMID: 37554279 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

