Cross-species complementation reveals conserved functions for EARLY FLOWERING 3 between monocots and dicots
- PMID: 31245666
- PMCID: PMC6508535
- DOI: 10.1002/pld3.18
Cross-species complementation reveals conserved functions for EARLY FLOWERING 3 between monocots and dicots
Abstract
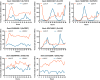
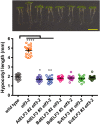
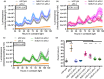
Plant responses to the environment are shaped by external stimuli and internal signaling pathways. In both the model plant Arabidopsis thaliana (Arabidopsis) and crop species, circadian clock factors are critical for growth, flowering, and circadian rhythms. Outside of Arabidopsis, however, little is known about the molecular function of clock gene products. Therefore, we sought to compare the function of Brachypodium distachyon (Brachypodium) and Setaria viridis (Setaria) orthologs of EARLY FLOWERING 3, a key clock gene in Arabidopsis. To identify both cycling genes and putative ELF3 functional orthologs in Setaria, a circadian RNA-seq dataset and online query tool (Diel Explorer) were generated to explore expression profiles of Setaria genes under circadian conditions. The function of ELF3 orthologs from Arabidopsis, Brachypodium, and Setaria was tested for complementation of an elf3 mutation in Arabidopsis. We find that both monocot orthologs were capable of rescuing hypocotyl elongation, flowering time, and arrhythmic clock phenotypes. Using affinity purification and mass spectrometry, our data indicate that BdELF3 and SvELF3 could be integrated into similar complexes in vivo as AtELF3. Thus, we find that, despite 180 million years of separation, BdELF3 and SvELF3 can functionally complement loss of ELF3 at the molecular and physiological level.
Keywords: ELF3; Setaria; circadian RNA‐seq; circadian clock; flowering; growth.
Figures


Similar articles
-
CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis.Plant Physiol. 2012 Feb;158(2):1079-88. doi: 10.1104/pp.111.189670. Epub 2011 Dec 21. Plant Physiol. 2012. PMID: 22190341 Free PMC article.
-
Gibberellin driven growth in elf3 mutants requires PIF4 and PIF5.Plant Signal Behav. 2015;10(3):e992707. doi: 10.4161/15592324.2014.992707. Plant Signal Behav. 2015. PMID: 25738547 Free PMC article.
-
Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA.Plant J. 2020 Mar;101(6):1397-1410. doi: 10.1111/tpj.14604. Epub 2019 Dec 11. Plant J. 2020. PMID: 31694066
-
Molecular and functional dissection of EARLY-FLOWERING 3 (ELF3) and ELF4 in Arabidopsis.Plant Sci. 2021 Feb;303:110786. doi: 10.1016/j.plantsci.2020.110786. Epub 2020 Dec 3. Plant Sci. 2021. PMID: 33487361 Review.
-
The intersection between circadian and heat-responsive regulatory networks controls plant responses to increasing temperatures.Biochem Soc Trans. 2022 Jun 30;50(3):1151-1165. doi: 10.1042/BST20190572. Biochem Soc Trans. 2022. PMID: 35758233 Free PMC article. Review.
Cited by
-
Genetic basis of phenotypic plasticity and genotype × environment interactions in a multi-parental tomato population.J Exp Bot. 2020 Sep 19;71(18):5365-5376. doi: 10.1093/jxb/eraa265. J Exp Bot. 2020. PMID: 32474596 Free PMC article.
-
Maize stigmas react differently to self- and cross-pollination and fungal invasion.Plant Physiol. 2024 Dec 2;196(4):3071-3090. doi: 10.1093/plphys/kiae536. Plant Physiol. 2024. PMID: 39371027 Free PMC article.
-
PHYTOCHROME C regulation of photoperiodic flowering via PHOTOPERIOD1 is mediated by EARLY FLOWERING 3 in Brachypodium distachyon.PLoS Genet. 2023 May 10;19(5):e1010706. doi: 10.1371/journal.pgen.1010706. eCollection 2023 May. PLoS Genet. 2023. PMID: 37163541 Free PMC article.
-
Phytochromes transmit photoperiod information via the evening complex in Brachypodium.Genome Biol. 2023 Nov 7;24(1):256. doi: 10.1186/s13059-023-03082-w. Genome Biol. 2023. PMID: 37936225 Free PMC article.
-
FaesAP3_1 Regulates the FaesELF3 Gene Involved in Filament-Length Determination of Long-Homostyle Fagopyrum esculentum.Int J Mol Sci. 2022 Nov 19;23(22):14403. doi: 10.3390/ijms232214403. Int J Mol Sci. 2022. PMID: 36430880 Free PMC article.
References
-
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. - PubMed
-
- Bendix, C. , Marshall, C. M. , & Harmon, F. G. (2015). Circadian clock genes universally control key agricultural traits. Molecular Plant, 8, 1135–1152. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

