Genomewide association study of ionomic traits on diverse soybean populations from germplasm collections
- PMID: 31245681
- PMCID: PMC6508489
- DOI: 10.1002/pld3.33
Genomewide association study of ionomic traits on diverse soybean populations from germplasm collections
Abstract
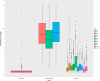
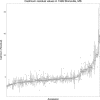
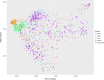
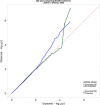
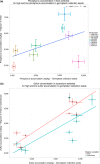
The elemental content of a soybean seed is a determined by both genetic and environmental factors and is an important component of its nutritional value. The elemental content is chemically stable, making the samples stored in germplasm repositories an intriguing source of experimental material. To test the efficacy of using samples from germplasm banks for gene discovery, we analyzed the elemental profile of seeds from 1,653 lines in the USDA Soybean Germplasm Collection. We observed large differences in the elemental profiles based on where the lines were grown, which lead us to break up the genetic analysis into multiple small experiments. Despite these challenges, we were able to identify candidate single nucleotide polymorphisms (SNPs) controlling elemental accumulation as well as lines with extreme elemental accumulation phenotypes. Our results suggest that elemental analysis of germplasm samples can identify SNPs in linkage disequilibrium to genes, which can be leveraged to assist in crop improvement efforts.
Keywords: GRIN; GWAS; diversity panel; ionomics; multi‐locus mixed model; soybean.
Figures
References
LinkOut - more resources
Full Text Sources

