Transcriptomic analysis of sweet potato under dehydration stress identifies candidate genes for drought tolerance
- PMID: 31245692
- PMCID: PMC6508841
- DOI: 10.1002/pld3.92
Transcriptomic analysis of sweet potato under dehydration stress identifies candidate genes for drought tolerance
Abstract
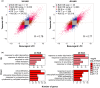
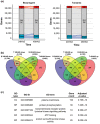
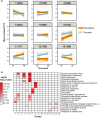
Sweet potato (Ipomoea batatas [L.] Lam.) is an important subsistence crop in Sub-Saharan Africa, yet as for many crops, yield can be severely impacted by drought stress. Understanding the genetic mechanisms that control drought tolerance can facilitate the development of drought-tolerant sweet potato cultivars. Here, we report an expression profiling study using the US-bred cultivar, Beauregard, and a Ugandan landrace, Tanzania, treated with polyethylene glycol (PEG) to simulate drought and sampled at 24 and 48 hr after stress. At each time-point, between 4,000 to 6,000 genes in leaf tissue were differentially expressed in each cultivar. Approximately half of these differentially expressed genes were common between the two cultivars and were enriched for Gene Ontology terms associated with drought response. Three hundred orthologs of drought tolerance genes reported in model species were identified in the Ipomoea trifida reference genome, of which 122 were differentially expressed under at least one experimental condition, constituting a list of drought tolerance candidate genes. A subset of genes was differentially regulated between Beauregard and Tanzania, representing genotype-specific responses to drought stress. The data analyzed and reported here provide a resource for geneticists and breeders toward identifying and utilizing drought tolerance genes in sweet potato.
Keywords: Ipomoea batatas; RNA‐Seq; drought; expression; polyethylene glycol; sweet potato.
Figures


Similar articles
-
Transcriptome Analysis Reveals Genes and Pathways Associated with Drought Tolerance of Early Stages in Sweet Potato (Ipomoea batatas (L.) Lam.).Genes (Basel). 2024 Jul 19;15(7):948. doi: 10.3390/genes15070948. Genes (Basel). 2024. PMID: 39062727 Free PMC article.
-
Flooding Tolerance in Sweet Potato (Ipomoea batatas (L.) Lam) Is Mediated by Reactive Oxygen Species and Nitric Oxide.Antioxidants (Basel). 2022 Apr 29;11(5):878. doi: 10.3390/antiox11050878. Antioxidants (Basel). 2022. PMID: 35624742 Free PMC article.
-
Transcriptomic analysis reveals mechanisms for the different drought tolerance of sweet potatoes.Front Plant Sci. 2023 Mar 16;14:1136709. doi: 10.3389/fpls.2023.1136709. eCollection 2023. Front Plant Sci. 2023. PMID: 37008495 Free PMC article.
-
Sweet Potato as a Key Crop for Food Security under the Conditions of Global Climate Change: A Review.Plants (Basel). 2023 Jun 30;12(13):2516. doi: 10.3390/plants12132516. Plants (Basel). 2023. PMID: 37447081 Free PMC article. Review.
-
Current status in whole genome sequencing and analysis of Ipomoea spp.Plant Cell Rep. 2019 Nov;38(11):1365-1371. doi: 10.1007/s00299-019-02464-4. Epub 2019 Aug 29. Plant Cell Rep. 2019. PMID: 31468128 Free PMC article. Review.
Cited by
-
Multiple QTL Mapping in Autopolyploids: A Random-Effect Model Approach with Application in a Hexaploid Sweetpotato Full-Sib Population.Genetics. 2020 Jul;215(3):579-595. doi: 10.1534/genetics.120.303080. Epub 2020 May 5. Genetics. 2020. PMID: 32371382 Free PMC article.
-
Identification of genes associated with abiotic stress tolerance in sweetpotato using weighted gene co-expression network analysis.Plant Direct. 2023 Oct 3;7(10):e532. doi: 10.1002/pld3.532. eCollection 2023 Oct. Plant Direct. 2023. PMID: 37794882 Free PMC article.
-
Quantitative trait loci and differential gene expression analyses reveal the genetic basis for negatively associated β-carotene and starch content in hexaploid sweetpotato [Ipomoea batatas (L.) Lam.].Theor Appl Genet. 2020 Jan;133(1):23-36. doi: 10.1007/s00122-019-03437-7. Epub 2019 Oct 8. Theor Appl Genet. 2020. PMID: 31595335 Free PMC article.
-
Mitogen-activated protein kinase 11 (MAPK11) maintains growth and photosynthesis of potato plant under drought condition.Plant Cell Rep. 2021 Mar;40(3):491-506. doi: 10.1007/s00299-020-02645-6. Epub 2021 Jan 3. Plant Cell Rep. 2021. PMID: 33388892
-
Transcriptome Analysis Reveals Genes and Pathways Associated with Drought Tolerance of Early Stages in Sweet Potato (Ipomoea batatas (L.) Lam.).Genes (Basel). 2024 Jul 19;15(7):948. doi: 10.3390/genes15070948. Genes (Basel). 2024. PMID: 39062727 Free PMC article.
References
-
- Aguirrezabal, L. , Bouchier‐Combaud, S. , Radziejwoski, A. , Dauzat, M. , Cookson, S. J. , & Granier, C. (2006). Plasticity to soil water deficit in Arabidopsis thaliana: Dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant, Cell and Environment, 29, 2216–2227. 10.1111/j.1365-3040.2006.01595.x - DOI - PubMed
-
- Alexa, A. , & Rahnenfuhrer, J. (2016). topGO: Enrichment Analysis for Gene Ontology.
-
- Austin, D. F. (1988). The taxonomy, evolution and genetic diversity of sweet potatoes and related wild species In Gregory P. (Ed.), Exploration, maintenance and utilization of sweet potato genetic resources, Report of the 1st Sweet Potato Planning Conference 1987 (pp. 27–59). Lima, Peru: International Potato Center.
LinkOut - more resources
Full Text Sources

