Development of an activation tagging system for maize
- PMID: 31245761
- PMCID: PMC6508757
- DOI: 10.1002/pld3.118
Development of an activation tagging system for maize
Abstract
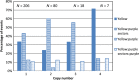
Activation Tagging, distributing transcriptional enhancers throughout the genome to induce transcription of nearby genes, is a powerful tool for discovering the function of genes in plants. We have developed a transposable element system to distribute a novel activation tagging element throughout the genome of maize. The transposon system is built from the Enhancer/Suppressor (En/Spm) transposon system and uses an engineered seed color marker to show when the transposon excises. Both somatic and germinal excision events can be detected by the seed color. The activation tagging element is in a Spm-derived non-autonomous transposon and contains four copies of the Sugarcane Bacilliform Virus-enhancer (SCBV-enhancer) and the AAD1 selectable marker. We have demonstrated that the transposon can give rise to germinal excision events that can re-integrate into non-linked genomic locations. The transposon has remained active for three generations and events displaying high rates of germinal excision in the T2 generation have been identified. This system can generate large numbers of activation tagged maize lines that can be screened for agriculturally relevant phenotypes.
Keywords: activation tagging; enhancer; insertional mutagenesis; transposable elements.
Figures








References
-
- Becraft, P. W. , & Yi, G. (2010). Regulation of aleurone development in cereal grains. Journal of Experimental Botany, 62, 1669–1675. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

