Defining Genetic Variation in Widely Used Congenic and Backcrossed Mouse Models Reveals Varied Regulation of Genes Important for Immune Responses
- PMID: 31248780
- PMCID: PMC6883924
- DOI: 10.1016/j.immuni.2019.05.006
Defining Genetic Variation in Widely Used Congenic and Backcrossed Mouse Models Reveals Varied Regulation of Genes Important for Immune Responses
Abstract
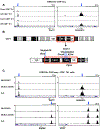
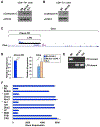
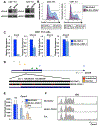
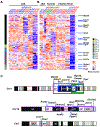
Genetic variation influences how the genome is interpreted in individuals and in mouse strains used to model immune responses. We developed approaches to utilize next-generation sequencing datasets to identify sequence variation in genes and enhancer elements in congenic and backcross mouse models. We defined genetic variation in the widely used B6-CD45.2 and B6.SJL-CD45.1 congenic model, identifying substantial differences in SJL genetic content retained in B6.SJL-CD45.1 strains on the basis of the vendor source of the mice. Genes encoding PD-1, CD62L, Bcl-2, cathepsin E, and Cxcr4 were within SJL genetic content in at least one vendor source of B6.SJL-CD45.1 mice. SJL genetic content affected enhancer elements, gene regulation, protein expression, and amino acid content in CD4+ T helper 1 cells, and mice infected with influenza showed reduced expression of Cxcr4 on B6.SJL-CD45.1 T follicular helper cells. These findings provide information on experimental variables and aid in creating approaches that account for genetic variables.
Keywords: CD45.1; CD8(+) T cells; Cxcr4; ERV; Ly5.1; Rgs16; T-bet; Tfh; Th1; germ-free.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Mouse Watch: A Cautionary Tale.Immunity. 2019 Jul 16;51(1):10-12. doi: 10.1016/j.immuni.2019.06.019. Immunity. 2019. PMID: 31315029
Similar articles
-
Congenic interval of CD45/Ly-5 congenic mice contains multiple genes that may influence hematopoietic stem cell engraftment.Blood. 2010 Jan 14;115(2):408-17. doi: 10.1182/blood-2008-03-143370. Epub 2009 Nov 9. Blood. 2010. PMID: 19901263 Free PMC article.
-
Generation of a C57BL/6J mouse strain expressing the CD45.1 epitope to improve hematopoietic stem cell engraftment and adoptive cell transfer experiments.Lab Anim (NY). 2023 Dec;52(12):324-331. doi: 10.1038/s41684-023-01275-1. Epub 2023 Nov 28. Lab Anim (NY). 2023. PMID: 38017180
-
Single Targeted Exon Mutation Creates a True Congenic Mouse for Competitive Hematopoietic Stem Cell Transplantation: The C57BL/6-CD45.1(STEM) Mouse.Stem Cell Reports. 2016 Jun 14;6(6):985-992. doi: 10.1016/j.stemcr.2016.04.010. Epub 2016 May 12. Stem Cell Reports. 2016. PMID: 27185283 Free PMC article.
-
Murine susceptibility to mercury. II. autoantibody profiles and renal immune deposits in hybrid, backcross, and H-2d congenic mice.Clin Immunol Immunopathol. 1993 Jul;68(1):9-20. doi: 10.1006/clin.1993.1088. Clin Immunol Immunopathol. 1993. PMID: 8513597
-
Tfh1 Cells in Germinal Centers During Chronic HIV/SIV Infection.Front Immunol. 2018 Jun 6;9:1272. doi: 10.3389/fimmu.2018.01272. eCollection 2018. Front Immunol. 2018. PMID: 29928280 Free PMC article. Review.
Cited by
-
Nanoparticle pre-treatment for enhancing the survival and activation of pulmonary macrophage transplant.Drug Deliv Transl Res. 2023 Jul;13(7):1955-1966. doi: 10.1007/s13346-023-01319-6. Epub 2023 Mar 14. Drug Deliv Transl Res. 2023. PMID: 36917409 Free PMC article.
-
Investigating the Role of Inflammasome Caspases 1 and 11 in the Acute Radiation Syndrome.Radiat Res. 2021 Dec 1;196(6):686-689. doi: 10.1667/RADE-21-00141.1. Radiat Res. 2021. PMID: 34644390 Free PMC article.
-
Inflammation switches the chemoattractant requirements for naive lymphocyte entry into lymph nodes.Cell. 2025 Feb 20;188(4):1019-1035.e22. doi: 10.1016/j.cell.2024.11.031. Epub 2024 Dec 20. Cell. 2025. PMID: 39708807 Free PMC article.
-
Anti-Mouse CD83 Monoclonal Antibody Targeting Mature Dendritic Cells Provides Protection Against Collagen Induced Arthritis.Front Immunol. 2022 Feb 10;13:784528. doi: 10.3389/fimmu.2022.784528. eCollection 2022. Front Immunol. 2022. PMID: 35222372 Free PMC article.
-
Combination blockade of KLRG1 and PD-1 promotes immune control of local and disseminated cancers.Oncoimmunology. 2021 Jun 15;10(1):1933808. doi: 10.1080/2162402X.2021.1933808. Oncoimmunology. 2021. PMID: 34188973 Free PMC article.
References
-
- Chisolm DA, Savic D, Moore AJ, Ballesteros-Tato A, Leon B, Crossman DK, Murre C, Myers RM, and Weinmann AS (2017). CCCTC-Binding Factor Translates Interleukin 2- and alpha-Ketoglutarate-Sensitive Metabolic Changes in T Cells into Context-Dependent Gene Programs. Immunity 47, 251–267 e257. - PMC - PubMed
-
- Chisolm DA, and Weinmann AS (2018). Connections Between Metabolism and Epigenetics in Programming Cellular Differentiation. Annu Rev Immunol 36, 221–246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

